7Y0Y
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA (SeMet) | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7Y0Z
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA | | Descriptor: | TetR family transcriptional regulator | | Authors: | Liang, H, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
8V1X
 
 | | Crystal structure of polyketide synthase (PKS) thioreductase domain from Streptomyces coelicolor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pereira, J.H, Dan, Q, Keasling, J, Adams, P.D. | | Deposit date: | 2023-11-21 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A polyketide-based biosynthetic platform for diols, amino alcohols and hydroxy acids
Nat Catal, 8, 2025
|
|
4H1M
 
 | | Crystal structure of PYK2 with the indole 10c | | Descriptor: | 7-({[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]carbamoyl}amino)-N-(propan-2-yl)-1H-indole-2-carboxamide, Protein-tyrosine kinase 2-beta | | Authors: | Han, S. | | Deposit date: | 2012-09-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of novel series of pyrazole and indole-urea based DFG-out PYK2 inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8HJ4
 
 | |
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
4HVA
 
 | |
4LCZ
 
 | | Crystal structure of a multilayer-packed major light-harvesting complex | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Wan, T, Li, M, Chang, W.R. | | Deposit date: | 2013-06-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a multilayer packed major light-harvesting complex: implications for grana stacking in higher plants.
Mol Plant, 7, 2014
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | Descriptor: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S},3~{R})-4-(cyclopentylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8XXT
 
 | | ASFV RNAP M1249L C-tail occupied complex2 (MCOC2) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XXP
 
 | | ASFV RNAP core complex | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XX4
 
 | | ASFV RNAP elongation complex | | Descriptor: | C122R, D339L, DNA (5'-D(P*CP*GP*CP*AP*AP*CP*GP*GP*CP*GP*A)-3'), ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8Y0E
 
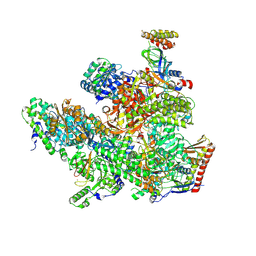 | | ASFV RNAP M1249L C-tail occupied complex4 (MCOC4) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XX5
 
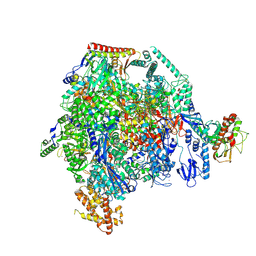 | | ASFV RNAP M1249L C-tail occupied complex1 (MCOC1) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XY6
 
 | | ASFV RNAP M1249L C-tail occupied complex3 (MCOC3) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8YZD
 
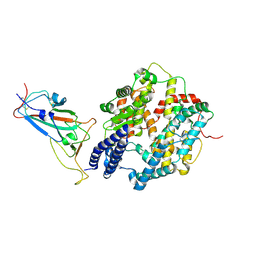 | | Structure of JN.1 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZE
 
 | | The JN.1 spike protein (S) in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZC
 
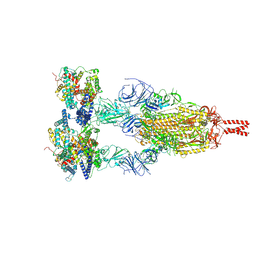 | | Structure of BA.2.86 spike protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZB
 
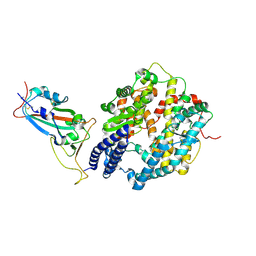 | | BA.2.86 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8KBE
 
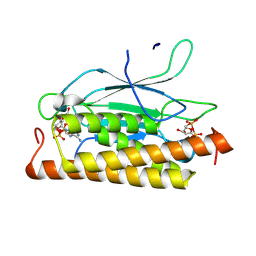 | | Structure of CbTad1 complexed with 1',3'-cADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Thoeris anti-defense 1 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBF
 
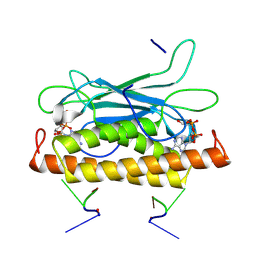 | | Structure of CbTad1 complexed with 1',3'-cADPR and cA3 | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, Thoeris anti-defense 1 | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBB
 
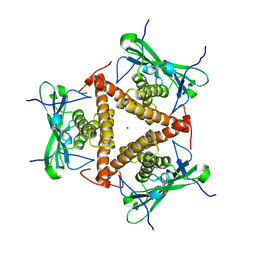 | | Structure of apo-CmTad1 | | Descriptor: | Thoeris anti-defense 1, ZINC ION | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
8KBL
 
 | | Structure of AcrIIA7 complexed with 1',3'-cADPR and cGG | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Inhibitor of Type II CRISPR-Cas system | | Authors: | Xiao, Y, Feng, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Single phage proteins sequester signals from TIR and cGAS-like enzymes.
Nature, 635, 2024
|
|
