5XSN
 
 | | The catalytic domain of GdpP with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XSP
 
 | | The catalytic domain of GdpP with 5'-pApA | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XT3
 
 | | The catalytic domain of GdpP with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5XSI
 
 | | The catalytic domain of GdpP | | Descriptor: | MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
5WJK
 
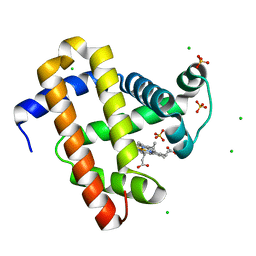 | | 2.0-Angstrom In situ Mylar structure of sperm whale myoglobin (SWMb) at 293 K | | Descriptor: | CHLORIDE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Broecker, J, Ou, W.-L, Ernst, O.P. | | Deposit date: | 2017-07-23 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
5WKT
 
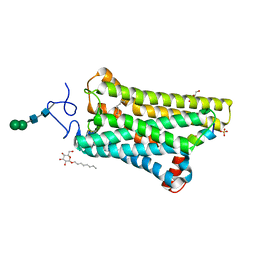 | | 3.2-Angstrom In situ Mylar structure of bovine opsin at 100 K | | Descriptor: | Rhodopsin, SULFATE ION, Transducin Galpha peptide, ... | | Authors: | Broecker, J, Morizumi, T, Ou, W.-L, Ernst, O.P. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions.
Nat Protoc, 13, 2018
|
|
6A06
 
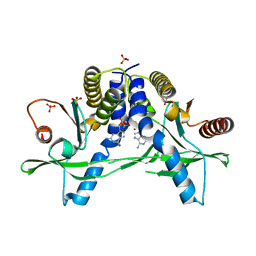 | | Structure of pSTING complex | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
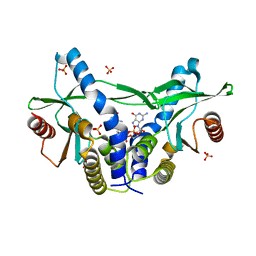 | | Structure of pSTING complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
8HQ6
 
 | | KL2 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8HQ3
 
 | | KL1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
8J4T
 
 | | GajA-GajB complex | | Descriptor: | Endonuclease GajA, Gabija protein GajB | | Authors: | Wei, T, Ma, J, Huo, Y. | | Deposit date: | 2023-04-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and biochemical insights into the mechanism of the Gabija bacterial immunity system.
Nat Commun, 15, 2024
|
|
7UU6
 
 | |
7UU8
 
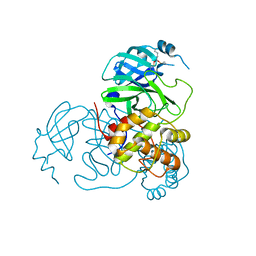 | |
7UUC
 
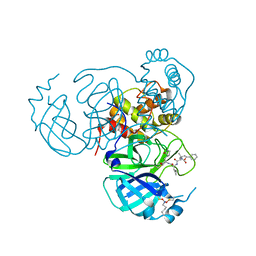 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | Descriptor: | 3C-like proteinase nsp5, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UU7
 
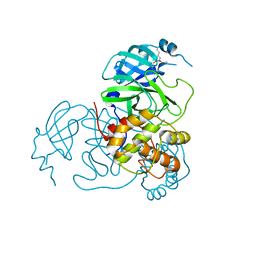 | |
7UUB
 
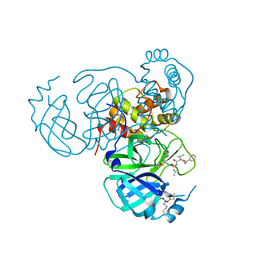 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI12 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UUA
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase nsp5, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UU9
 
 | |
7UUE
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI85 | | Descriptor: | 3C-like proteinase nsp5, benzyl [(2S,3R)-1-({(2S)-1-[2-(3-amino-3-oxopropyl)-2-propanoylhydrazinyl]-3-cyclohexyl-1-oxopropan-2-yl}amino)-3-tert-butoxy-1-oxobutan-2-yl]carbamate (non-preferred name) | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7UUD
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7VFU
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to ziconotide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFV
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to PD173212 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-(~{tert}-butylamino)-1-oxidanylidene-3-(4-phenylmethoxyphenyl)propan-2-yl]-2-[(4-~{tert}-butylphenyl)methyl-methyl-amino]-4-methyl-pentanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFW
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, bound to CaV2.2-blocker1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(4-chlorophenyl)-1-(2-methoxyphenyl)-3-(2,2,6,6-tetramethyloxan-4-yl)pyrazole, CALCIUM ION, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VFS
 
 | | Human N-type voltage gated calcium channel CaV2.2-alpha2/delta1-beta1 complex, apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dong, Y, Gao, Y, Wang, Y, Zhao, Y. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Closed-state inactivation and pore-blocker modulation mechanisms of human Ca V 2.2.
Cell Rep, 37, 2021
|
|
7VLN
 
 | | NSD2-PWWP1 domain bound with an imidazol-5-yl benzonitrile compound | | Descriptor: | 4-[5-[4-(aminomethyl)-2,6-dimethoxy-phenyl]-3-methyl-imidazol-4-yl]benzenecarbonitrile, Histone-lysine N-methyltransferase NSD2 | | Authors: | Cao, D.Y, Li, Y.L, Li, J, Xiong, B. | | Deposit date: | 2021-10-05 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Based Discovery of a Series of NSD2-PWWP1 Inhibitors.
J.Med.Chem., 65, 2022
|
|
