7L9M
 
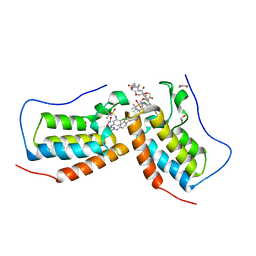 | | Crystal structure of the first bromodomain (BD1) of human BRD4 in complex with bivalent inhibitor GXH-II-083 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-[(1,19-dioxo-4,7,10,13,16-pentaoxanonadecane-1,19-diyl)di(piperidine-1,4-diyl)]bis(4-{[4-({3-[(tert-butylsulfonyl)amino]-4-chlorophenyl}amino)-5-methylpyrimidin-2-yl]amino}-2-fluorobenzamide) | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
7LL4
 
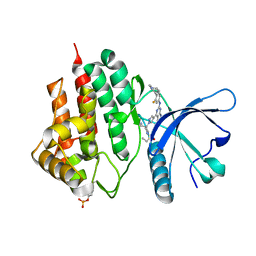 | |
7LB3
 
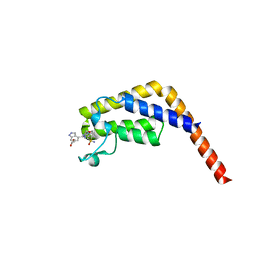 | |
7LL5
 
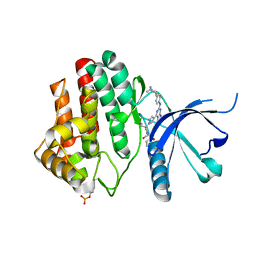 | |
7LP0
 
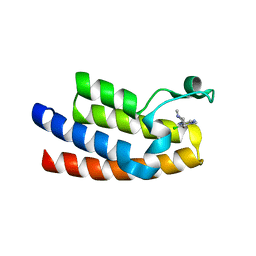 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-077 | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorol-2-methyl-5-[[(3~{R})-1-methylpiperidin-3-yl]amino]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LRK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor Pdy-3-093 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{S})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LRO
 
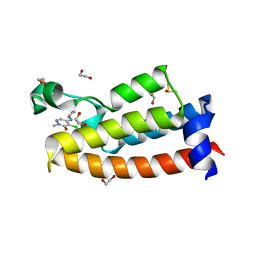 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-01-105 | | Descriptor: | 1,2-ETHANEDIOL, 5-(azetidin-3-ylamino)-4-chloranyl-2-methyl-pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-17 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7LPK
 
 | | Crystal structure of BPTF bromodomain in complex with inhibitor HZ-03-112 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-2-methyl-5-[[(3~{R})-pyrrolidin-3-yl]amino]pyridazin-3-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-02-12 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7L9G
 
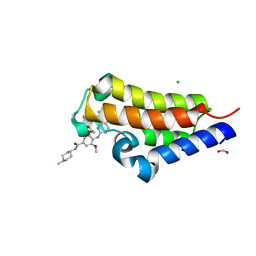 | | Crystal structure of the second bromodomain (BD2) of human BRD2 bound to BI2536 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain-containing protein 2, ... | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
7LA9
 
 | |
7N42
 
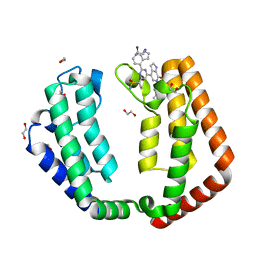 | | Crystal structure of the tandem bromodomain of human TAF1 (TAF1-T) bound to ZS1-681 | | Descriptor: | 1,2-ETHANEDIOL, 6-(but-3-en-1-yl)-4-[6-{1-[(R)-S-methanesulfonimidoyl]cyclopropyl}-2-(1H-pyrrolo[2,3-b]pyridin-4-yl)pyrimidin-4-yl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, DIMETHYL SULFOXIDE, ... | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2021-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tandem bromodomain of human TAF1 (TAF1-T) bound to ZS1-681
To Be Published
|
|
7MR9
 
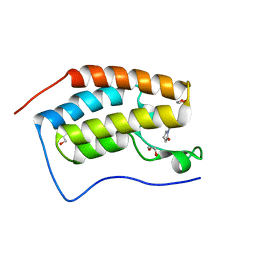 | |
7MRA
 
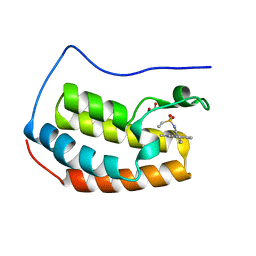 | |
7MRB
 
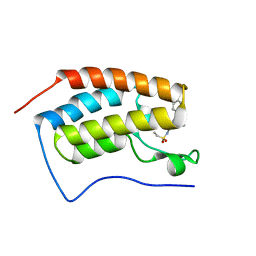 | |
7MRH
 
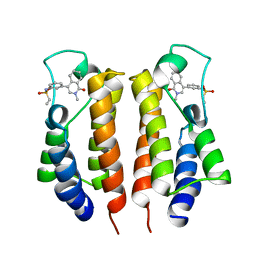 | |
7MRG
 
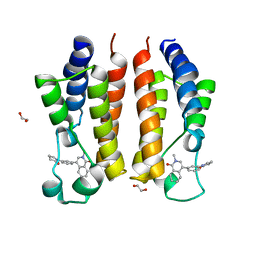 | |
7MPH
 
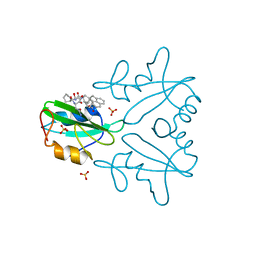 | | GRB2 SH2 Domain with Compound 7 | | Descriptor: | (4-{(10R,11E,14S,18S)-18-(2-amino-2-oxoethyl)-14-[(naphthalen-1-yl)methyl]-8,17,20-trioxo-7,16,19-triazaspiro[5.14]icos-11-en-10-yl}phenyl)acetic acid, 1,2-ETHANEDIOL, Growth factor receptor-bound protein 2, ... | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-05-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structural characterization of a monocarboxylic inhibitor for GRB2 SH2 domain.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
7MR7
 
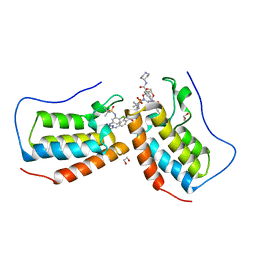 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-075 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluoro-N-[1-(14-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]phenyl}-14-oxo-4,7,10-trioxa-13-azatetradecanan-1-oyl)piperidin-4-yl]benzamide, Bromodomain-containing protein 4 | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
7MR8
 
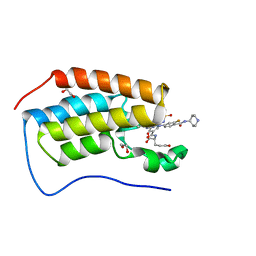 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-076 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-{3-[(2-{3-fluoro-4-[(piperidin-4-yl)carbamoyl]anilino}-5-methylpyrimidin-4-yl)amino]-5-[(2-methylpropane-2-sulfonyl)amino]benzoyl}-L-glutamic acid | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-076
To Be Published
|
|
7MR5
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-052 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-{oxybis[(ethane-2,1-diyl)oxy(1-oxoethane-2,1-diyl)piperidine-1,4-diyl]}bis{4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluorobenzamide} | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
7MRC
 
 | |
7MR6
 
 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to GXH-II-082 | | Descriptor: | Bromodomain-containing protein 4, N,N'-[(1,16-dioxo-4,7,10,13-tetraoxahexadecane-1,16-diyl)di(piperidine-1,4-diyl)]bis{4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluorobenzamide} | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
7MRD
 
 | | Crystal structure of the first bromodomain (BD1) of human BRDT bound to GXH-II-082 | | Descriptor: | Bromodomain testis-specific protein, N,N'-[(1,16-dioxo-4,7,10,13-tetraoxahexadecane-1,16-diyl)di(piperidine-1,4-diyl)]bis{4-[(4-{4-chloro-3-[(2-methylpropane-2-sulfonyl)amino]anilino}-5-methylpyrimidin-2-yl)amino]-2-fluorobenzamide} | | Authors: | Chan, A, Schonbrunn, E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Bivalent BET Bromodomain Inhibitors Confer Increased Potency and Selectivity for BRDT via Protein Conformational Plasticity.
J.Med.Chem., 65, 2022
|
|
3NY1
 
 | | Structure of the ubr-box of the UBR1 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
