1V92
 
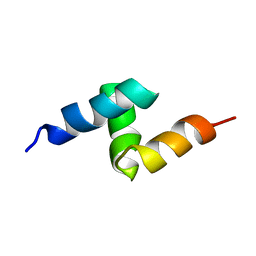 | | Solution structure of the UBA domain from p47, a major cofactor of the AAA ATPase p97 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-01-19 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97
Embo J., 23, 2004
|
|
1T28
 
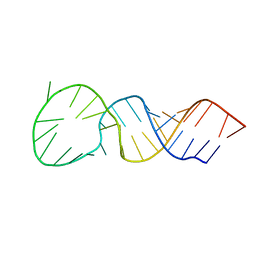 | | High resolution structure of a picornaviral internal cis-acting replication element | | Descriptor: | 34-MER | | Authors: | Thiviyanathan, V, Yang, Y, Kaluarachchi, K, Reynbrand, R, Gorenstein, D.G, Lemon, S.M. | | Deposit date: | 2004-04-20 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of a picornaviral internal cis-acting replication element(cre).
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3WVE
 
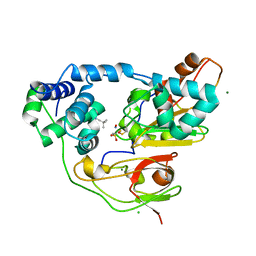 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, before photo-activation | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-05-17 | | Release date: | 2015-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1WZK
 
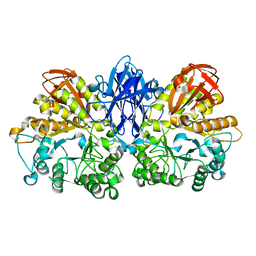 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt D465N | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1WZL
 
 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469L | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
3X25
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 700 min | | Descriptor: | 2,2-dimethylpropanenitrile, CHLORIDE ION, FE (III) ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3X24
 
 | | Crystal structure of Nitrile Hydratase mutant bR56K complexed with Trimethylacetonitrile, photo-activated for 120 min | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2014-12-10 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Time-Resolved Crystallography of the Reaction Intermediate of Nitrile Hydratase: Revealing a Role for the Cysteinesulfenic Acid Ligand as a Catalytic Nucleophile.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1IYS
 
 | | Crystal Structure of Class A beta-Lactamase Toho-1 | | Descriptor: | BETA-LACTAMASE TOHO-1, SULFATE ION | | Authors: | Ibuka, A.S, Ishii, Y, Yamaguchi, K, Matsuzawa, H, Sakai, H. | | Deposit date: | 2002-09-06 | | Release date: | 2003-10-14 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Extended-Spectrum beta-Lactamase Toho-1: Insights into the Molecular Mechanism for
Catalytic Reaction and Substrate Specificity Expansion
Biochemistry, 42, 2003
|
|
1WZM
 
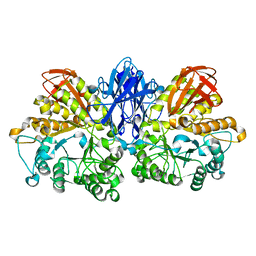 | | Thermoactinomyces vulgaris R-47 alpha-amylase II (TVA II) mutatnt R469K | | Descriptor: | Alpha-amylase II, CALCIUM ION | | Authors: | Mizuno, M, Ichikawa, K, Tonozuka, T, Ohtaki, A, Shimura, Y, Kamitori, S, Nishikawa, A, Sakano, Y. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mutagenesis and Structural Analysis of Thermoactinomyces vulgaris R-47 alpha-Amylase II (TVA II)
To be Published
|
|
1HQ0
 
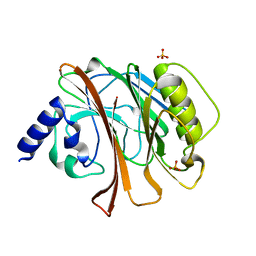 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF E.COLI CYTOTOXIC NECROTIZING FACTOR TYPE 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2000-12-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
1HZG
 
 | | CRYSTAL STRUCTURE OF THE INACTIVE C866S MUTANT OF THE CATALYTIC DOMAIN OF E. COLI CYTOTOXIC NECROTIZING FACTOR 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2001-01-24 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
1IHF
 
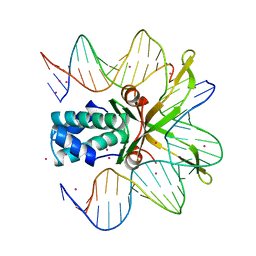 | | INTEGRATION HOST FACTOR/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (35-MER), DNA (5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*C P*AP*CP*C)-3'), ... | | Authors: | Rice, P.A, Yang, S.-W, Mizuuchi, K, Nash, H.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn.
Cell(Cambridge,Mass.), 87, 1996
|
|
6HBW
 
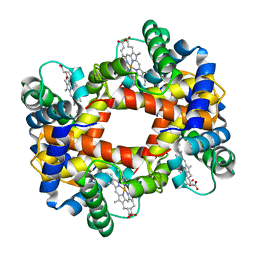 | |
1GBS
 
 | |
2XO8
 
 | | Crystal Structure of Myosin-2 in Complex with Tribromodichloropseudilin | | Descriptor: | 2,4-DICHLORO-6-(3,4,5-TRIBROMO-1H-PYRROL-2-YL)PHENOL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Preller, M, Chinthalapudi, K, Martin, R, Knoelker, H.J, Manstein, D.J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of Myosin ATPase Activity by Halogenated Pseudilins: A Structure-Activity Study.
J.Med.Chem., 54, 2011
|
|
2ONC
 
 | | Crystal structure of human DPP-4 | | Descriptor: | 2-({2-[(3R)-3-AMINOPIPERIDIN-1-YL]-4-OXOQUINAZOLIN-3(4H)-YL}METHYL)BENZONITRILE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Feng, J, Zhang, Z, Wallace, M.B, Stafford, J.A, Kaldor, S.W, Kassel, D.B, Navre, M, Shi, L, Skene, R.J, Asakawa, T, Takeuchi, K, Xu, R, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2007-01-23 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV.
J.Med.Chem., 50, 2007
|
|
2PDE
 
 | | THE HIGH RESOLUTION STRUCTURE OF THE PERIPHERAL SUBUNIT-BINDING DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM THE PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX OF BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Kalia, Y.N, Brocklehurst, S.M, Hipps, D.S, Appella, E, Sakaguchi, K, Perham, R.N. | | Deposit date: | 1992-11-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus.
J.Mol.Biol., 230, 1993
|
|
2FZI
 
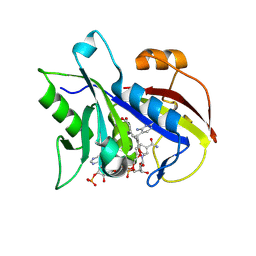 | | New Insights into DHFR Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[3',4'-DIMETHOXY-5'-(5-CARBOXYL-1-PENTYNYL)]BENZYL PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
2FZH
 
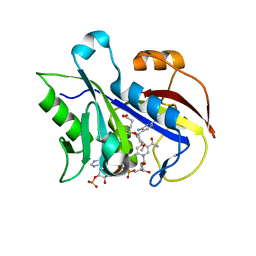 | | New Insights into Dihydrofolate Reductase Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
2FZJ
 
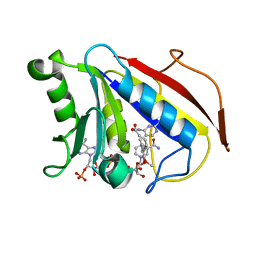 | | New Insights into DHFR Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[3',4'-DIMETHOXY-5'-(5-CARBOXYL-1-PENTYNYL)]BENZYL PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
1BQF
 
 | | GROWTH-BLOCKING PEPTIDE (GBP) FROM PSEUDALETIA SEPARATA | | Descriptor: | PROTEIN (GROWTH-BLOCKING PEPTIDE) | | Authors: | Aizawa, T, Fujitani, N, Hayakawa, Y, Ohnishi, A, Ohkubo, T, Kwano, K, Hikichi, K, Nitta, K. | | Deposit date: | 1998-08-09 | | Release date: | 1998-12-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an insect growth factor, growth-blocking peptide.
J.Biol.Chem., 274, 1999
|
|
2B86
 
 | |
2PDD
 
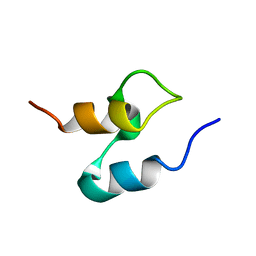 | | THE HIGH RESOLUTION STRUCTURE OF THE PERIPHERAL SUBUNIT-BINDING DOMAIN OF DIHYDROLIPOAMIDE ACETYLTRANSFERASE FROM THE PYRUVATE DEHYDROGENASE MULTIENZYME COMPLEX OF BACILLUS STEAROTHERMOPHILUS | | Descriptor: | DIHYDROLIPOAMIDE ACETYLTRANSFERASE | | Authors: | Kalia, Y.N, Brocklehurst, S.M, Hipps, D.S, Appella, E, Sakaguchi, K, Perham, R.N. | | Deposit date: | 1992-11-25 | | Release date: | 1994-12-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus.
J.Mol.Biol., 230, 1993
|
|
3VH5
 
 | | Crystal structure of the chicken CENP-T histone fold/CENP-W/CENP-S/CENP-X heterotetrameric complex, crystal form I | | Descriptor: | CENP-S, CENP-T, CENP-W, ... | | Authors: | Nishino, T, Takeuchi, K, Gascoigne, K.E, Suzuki, A, Hori, T, Oyama, T, Morikawa, K, Cheeseman, I.M, Fukagawa, T. | | Deposit date: | 2011-08-23 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | CENP-T-W-S-X Forms a Unique Centromeric Chromatin Structure with a Histone-like Fold
Cell(Cambridge,Mass.), 148, 2012
|
|
3WN8
 
 | | Crystal Structure of Collagen-Model Peptide, (POG)3-PRG-(POG)4 | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Haga, M, Noguchi, K, Tanaka, T. | | Deposit date: | 2013-12-06 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Preferred side-chain conformation of arginine residues in a triple-helical structure.
Biopolymers, 101, 2014
|
|
