6TEX
 
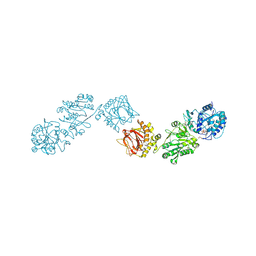 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Val80Lys mutant - Cocrystal with Fe2+, Mn2+, UDP-Glucose | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chiapparino, A, De Giorgi, F, Scietti, L, Faravelli, S, Roscioli, T, Forneris, F. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Regulatory Molecular 'Hot Spots' for LH/PLOD Collagen Glycosyltransferase Activity
Int J Mol Sci, 2023
|
|
7C2L
 
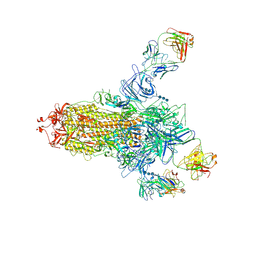 | | S protein of SARS-CoV-2 in complex bound with 4A8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Guo, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-05-08 | | Release date: | 2020-07-01 | | Last modified: | 2021-02-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2.
Science, 369, 2020
|
|
2KBR
 
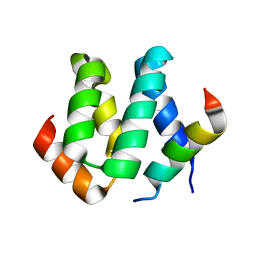 | | Solution structure of harmonin N terminal domain in complex with a internal peptide of cadherin23 | | Descriptor: | 18-meric peptide from Cadherin-23, Harmonin | | Authors: | Pan, L, Yan, J, Wu, L, Zhang, M. | | Deposit date: | 2008-12-05 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Assembling stable hair cell tip link complex via multidentate interactions between harmonin and cadherin 23
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6TAA
 
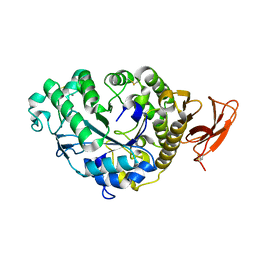 | | STRUCTURE AND MOLECULAR MODEL REFINEMENT OF ASPERGILLUS ORYZAE (TAKA) ALPHA-AMYLASE: AN APPLICATION OF THE SIMULATED-ANNEALING METHOD | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION | | Authors: | Swift, H.J, Brady, L, Derewenda, Z.S, Dodson, E.J, Turkenburg, J.P, Wilkinson, A.J. | | Deposit date: | 1992-08-21 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and molecular model refinement of Aspergillus oryzae (TAKA) alpha-amylase: an application of the simulated-annealing method.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
7AXU
 
 | |
2CGR
 
 | |
6JV3
 
 | |
7AXQ
 
 | |
7AXX
 
 | |
7AXP
 
 | |
2CL3
 
 | | Crystal structure of human Cleavage and Polyadenylation Specificity Factor 5 (CPSF5) | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 5 | | Authors: | Stenmark, P, Hogbom, M, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Nordlund, P. | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Human Cleavage and Polyadenylation Specific Factor-5 Reveals a Dimeric Nudix Protein with a Conserved Catalytic Site.
Proteins, 73, 2008
|
|
5JBF
 
 | | 4,6-alpha-glucanotransferase GTFB (D1015N mutant) from Lactobacillus reuteri 121 complexed with maltopentaose | | Descriptor: | CALCIUM ION, Inactive glucansucrase, SULFATE ION, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
3HLR
 
 | | Donor strand complemented FaeG of F4ad fimbriae | | Descriptor: | K88 fimbrial protein AD | | Authors: | Van Molle, I, Moonens, K, Garcia-Pino, A, Buts, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2009-05-28 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic characterization of pre- and postpolymerization states in the F4 fimbrial subunit FaeG
J.Mol.Biol., 394, 2009
|
|
6JV5
 
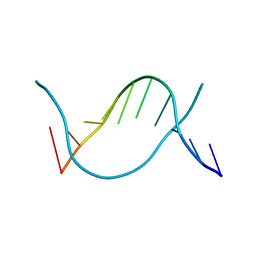 | | Crystal structure of 5-methylcytosine containing decamer dsDNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*(5CM)P*GP*CP*TP*GP*G)-3') | | Authors: | Zhang, L, Wang, Y.X. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Thymine DNA glycosylase recognizes the geometry alteration of minor grooves induced by 5-formylcytosine and 5-carboxylcytosine.
Chem Sci, 10, 2019
|
|
2I4R
 
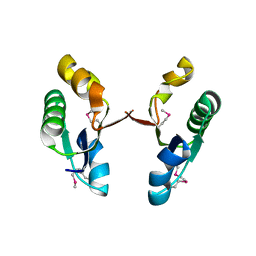 | | Crystal structure of the V-type ATP synthase subunit F from Archaeoglobus fulgidus. NESG target GR52A. | | Descriptor: | V-type ATP synthase subunit F | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Zhao, L, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-22 | | Release date: | 2006-08-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the V-type ATP synthase subunit F from Archaeoglobus fulgidus
To be Published
|
|
3HM6
 
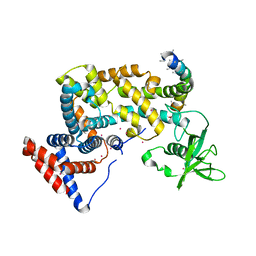 | | Crystal structure of the cytoplasmic domain of human plexin B1 | | Descriptor: | Plexin-B1, UNKNOWN ATOM OR ION, Unknown peptide | | Authors: | Tong, Y, He, H, Shen, L, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structure and function of the intracellular region of the plexin-b1 transmembrane receptor.
J.Biol.Chem., 284, 2009
|
|
1ZIM
 
 | |
1ZIL
 
 | |
1ZIK
 
 | |
4TGF
 
 | | SOLUTION STRUCTURES OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA DERIVED FROM 1*H NMR DATA | | Descriptor: | DES-VAL-1,VAL-2,TRANSFORMING GROWTH FACTOR, ALPHA | | Authors: | Kline, T.P, Brown, F.K, Brown, S.C, Jeffs, P.W, Kopple, K.D, Mueller, L. | | Deposit date: | 1990-06-13 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human transforming growth factor alpha derived from 1H NMR data.
Biochemistry, 29, 1990
|
|
3JAX
 
 | | Heavy meromyosin from Schistosoma mansoni muscle thick filament by negative stain EM | | Descriptor: | myosin 2 heavy chain, myosin regulatory light chain, smooth muscle myosin essential light chain | | Authors: | Sulbaran, G, Alamo, L, Pinto, A, Marquez, G, Mendez, F, Padron, R, Craig, R. | | Deposit date: | 2015-07-03 | | Release date: | 2015-10-07 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | An invertebrate smooth muscle with striated muscle myosin filaments.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6J9D
 
 | | Babesia microti lactate dehydrogenase R99A (BmLDHR99A) | | Descriptor: | L-lactate dehydrogenase | | Authors: | Yu, L. | | Deposit date: | 2019-01-22 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Crystal structures ofBabesia microtilactate dehydrogenase BmLDH reveal a critical role for Arg99 in catalysis.
Faseb J., 33, 2019
|
|
