4H37
 
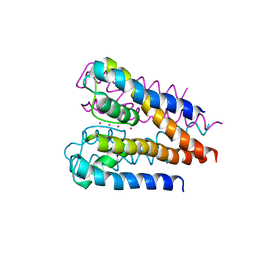 | | Crystal structure of a voltage-gated K+ channel pore domain in a closed state in lipid membranes | | Descriptor: | Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
3S8G
 
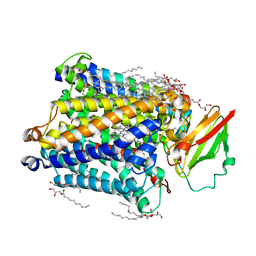 | | 1.8 A structure of ba3 cytochrome c oxidase mutant (A120F) from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
3S8F
 
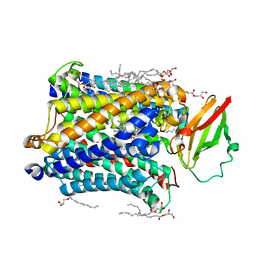 | | 1.8 A structure of ba3 cytochrome c oxidase from Thermus thermophilus in lipid environment | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Tiefenbrunn, T, Liu, W, Chen, Y, Katritch, V, Stout, C.D, Fee, J.A, Cherezov, V. | | Deposit date: | 2011-05-27 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structure of the ba3 cytochrome c oxidase from Thermus thermophilus in a lipidic environment.
Plos One, 6, 2011
|
|
2GLL
 
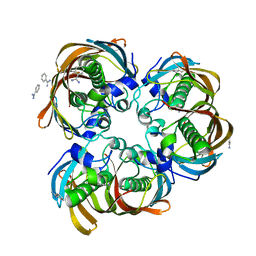 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
4H33
 
 | | Crystal structure of a voltage-gated K+ channel pore module in a closed state in lipid membranes, tetragonal crystal form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lmo2059 protein, POTASSIUM ION | | Authors: | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4IAQ
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4IB4
 
 | | Crystal structure of the chimeric protein of 5-HT2B-BRIL in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, ... | | Authors: | Wacker, D, Wang, C, Katritch, V, Han, G.W, Huang, X, Vardy, E, McCorvy, J.D, Jiang, Y, Chu, M, Siu, F.Y, Liu, W, Xu, H.E, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural features for functional selectivity at serotonin receptors.
Science, 340, 2013
|
|
2H1Y
 
 | |
2GLM
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with Compound 2 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 2-CHLORO-5-(5-{(E)-[(2Z)-3-(2-METHOXYETHYL)-4-OXO-2-(PHENYLIMINO)-1,3-THIAZOLIDIN-5-YLIDENE]METHYL}-2-FURYL)BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
4JKV
 
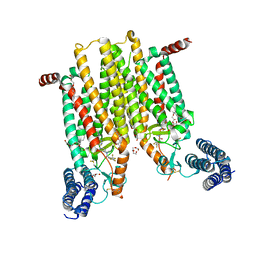 | | Structure of the human smoothened 7TM receptor in complex with an antitumor agent | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-fluoro-N-methyl-N-{1-[4-(1-methyl-1H-pyrazol-5-yl)phthalazin-1-yl]piperidin-4-yl}-2-(trifluoromethyl)benzamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, C, Wu, H, Katritch, V, Han, G.W, Huang, X, Liu, W, Siu, F.Y, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-03-11 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the human smoothened receptor bound to an antitumour agent.
Nature, 497, 2013
|
|
8I8F
 
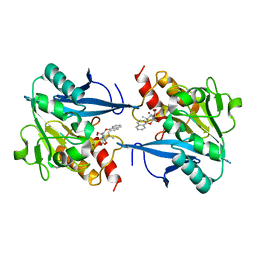 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed compound 1 | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-(2-naphthalen-1-yloxyethanoylamino)-2-oxidanyl-2-oxidanylidene-ethyl]-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, ZINC ION | | Authors: | Shi, X, Liu, W. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8F7W
 
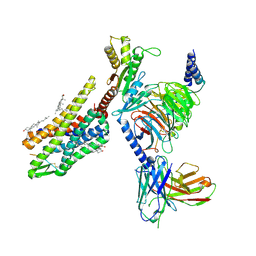 | | Gi bound kappa-opioid receptor in complex with dynorphin | | Descriptor: | CHOLESTEROL, Dynorphin, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Zhuang, Y, DiBerto, J.F, Zhou, X.E, Schmitz, G.P, Yuan, Q, Jain, M.K, Liu, W, Melcher, K, Jiang, Y, Roth, B.L, Xu, H.E. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structures of the entire human opioid receptor family.
Cell, 186, 2023
|
|
8F7R
 
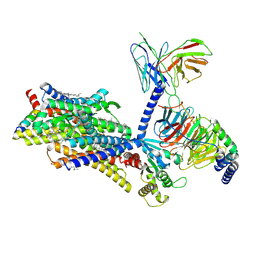 | | Gi bound mu-opioid receptor in complex with endomorphin | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, Y, Zhuang, Y, DiBerto, J.F, Zhou, X.E, Schmitz, G.P, Yuan, Q, Jain, M.K, Liu, W, Melcher, K, Jiang, Y, Roth, B.L, Xu, H.E. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structures of the entire human opioid receptor family.
Cell, 186, 2023
|
|
8F7Q
 
 | | Gi bound mu-opioid receptor in complex with beta-endorphin | | Descriptor: | Beta-endorphin, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Zhuang, Y, DiBerto, J.F, Zhou, X.E, Schmitz, G.P, Yuan, Q, Jain, M.K, Liu, W, Melcher, K, Jiang, Y, Roth, B.L, Xu, H.E. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structures of the entire human opioid receptor family.
Cell, 186, 2023
|
|
8F7S
 
 | | Gi bound delta-opioid receptor in complex with deltorphin | | Descriptor: | CHOLESTEROL, Delta-type opioid receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, Y, Zhuang, Y, DiBerto, J.F, Zhou, X.E, Schmitz, G.P, Yuan, Q, Jain, M.K, Liu, W, Melcher, K, Jiang, Y, Roth, B.L, Xu, H.E. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the entire human opioid receptor family.
Cell, 186, 2023
|
|
8F7X
 
 | | Gi bound nociceptin receptor in complex with nociceptin peptide | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Wang, Y, Zhuang, Y, DiBerto, J.F, Zhou, X.E, Schmitz, G.P, Yuan, Q, Jain, M.K, Liu, W, Melcher, K, Jiang, Y, Roth, B.L, Xu, H.E. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structures of the entire human opioid receptor family.
Cell, 186, 2023
|
|
8WAN
 
 | | Structure of transcribing complex 4 (TC4), the initially transcribing complex with Pol II positioned 4nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.07 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAS
 
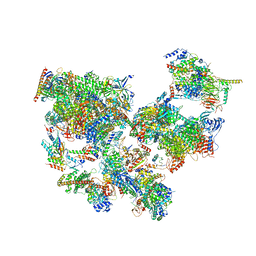 | | Structure of transcribing complex 9 (TC9), the initially transcribing complex with Pol II positioned 9nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (6.13 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAV
 
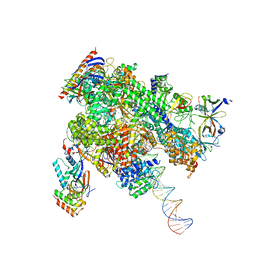 | | De novo transcribing complex 12 (TC12), the early elongation complex with Pol II positioned 12nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAP
 
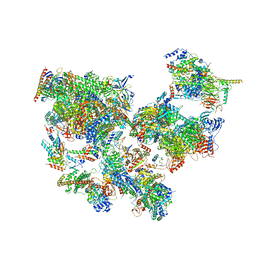 | | Structure of transcribing complex 6 (TC6), the initially transcribing complex with Pol II positioned 6nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (5.85 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAL
 
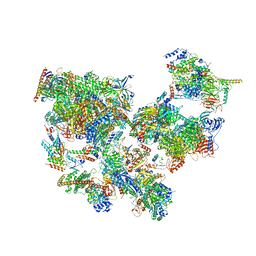 | | Structure of transcribing complex 3 (TC3), the initially transcribing complex with Pol II positioned 3nt downstream of TSS. | | Descriptor: | Alpha-amanitin, CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (8.52 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
8WAZ
 
 | | De novo transcribing complex 16 (TC16), the early elongation complex with Pol II positioned 16nt downstream of TSS | | Descriptor: | Alpha-amanitin, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Chen, X, Liu, W, Wang, Q, Wang, X, Ren, Y, Qu, X, Li, W, Xu, Y. | | Deposit date: | 2023-09-08 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural visualization of transcription initiation in action.
Science, 382, 2023
|
|
7X3O
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054 | | Descriptor: | (2~{R})-2-(3-fluoranyl-4-pyrimidin-5-yl-phenyl)butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07054
To Be Published
|
|
4L6R
 
 | | Structure of the class B human glucagon G protein coupled receptor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Soluble cytochrome b562 and Glucagon receptor chimera | | Authors: | Siu, F.Y, He, M, de Graaf, C, Han, G.W, Yang, D, Zhang, Z, Zhou, C, Xu, Q, Wacker, D, Joseph, J.S, Liu, W, Lau, J, Cherezov, V, Katritch, V, Wang, M.W, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the human glucagon class B G-protein-coupled receptor.
Nature, 499, 2013
|
|
