3HYK
 
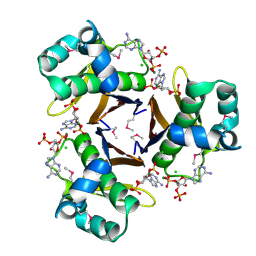 | | 2.31 Angstrom resolution crystal structure of a holo-(acyl-carrier-protein) synthase from Bacillus anthracis str. Ames in complex with CoA (3',5'-ADP) | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, CHLORIDE ION, Holo-[acyl-carrier-protein] synthase, ... | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-22 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2QUJ
 
 | |
4IS4
 
 | |
2QUH
 
 | |
2QUI
 
 | |
8AYV
 
 | |
4B2T
 
 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT ALPHA, T-COMPLEX PROTEIN 1 SUBUNIT BETA, T-COMPLEX PROTEIN 1 SUBUNIT DELTA, ... | | Authors: | Kalisman, N, Schroeder, G.F, Levitt, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
2X12
 
 | |
6NE7
 
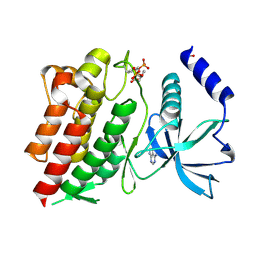 | | Structure of G810A mutant of RET protein tyrosine kinase domain. | | Descriptor: | ADENOSINE MONOPHOSPHATE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6NEC
 
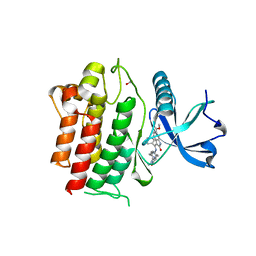 | | STRUCTURE OF RET PROTEIN TYROSINE KINASE DOMAIN IN COMPLEX WITH NINTEDANIB | | Descriptor: | FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret, methyl (3Z)-3-{[(4-{methyl[(4-methylpiperazin-1-yl)acetyl]amino}phenyl)amino](phenyl)methylidene}-2-oxo-2,3-dihydro-1H-indole-6-carboxylate | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2018-12-17 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
6NJA
 
 | | Structure of WT RET protein tyrosine kinase domain at 1.92A resolution. | | Descriptor: | ADENINE, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Terzyan, S.S, Shen, T, Wu, J, Mooers, B.H.M. | | Deposit date: | 2019-01-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of resistance of mutant RET protein-tyrosine kinase to its inhibitors nintedanib and vandetanib.
J.Biol.Chem., 294, 2019
|
|
2MJV
 
 | |
6OQA
 
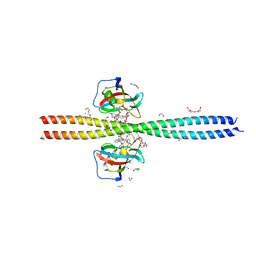 | | Crystal structure of CEP250 bound to FKBP12 in the presence of FK506-like novel natural product | | Descriptor: | (3R,4E,7E,10R,11S,12R,13S,16R,17R,24aS)-11,17-dihydroxy-10,12,16-trimethyl-3-[(2R)-1-phenylbutan-2-yl]-6,9,10,11,12,13,14,15,16,17,22,23,24,24a-tetradecahydro-3H-13,17-epoxypyrido[2,1-c][1,4]oxazacyclohenicosine-1,18,19(21H)-trione, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Lee, S.-J, Shigdel, U.K, Townson, S.A, Verdine, G.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5TCZ
 
 | |
1ECP
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-13 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Escherichia coli purine nucleoside phosphorylase: a comparison with the human enzyme reveals a conserved topology.
Structure, 5, 1997
|
|
4JM7
 
 | | 1.82 Angstrom resolution crystal structure of holo-(acyl-carrier-protein) synthase (acpS) from Staphylococcus aureus | | Descriptor: | Holo-[acyl-carrier-protein] synthase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Structural characterization and comparison of three acyl-carrier-protein synthases from pathogenic bacteria.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4L1W
 
 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
4L1X
 
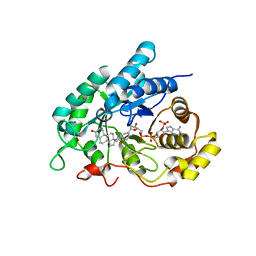 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 V54L Mutant in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
5N3U
 
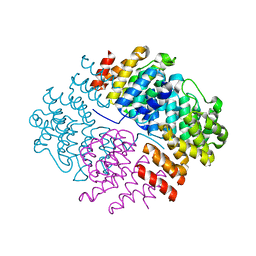 | | The structure of the complex of CpcE and CpcF of phycocyanin lyase from Nostoc sp. PCC7120 | | Descriptor: | Phycocyanobilin lyase subunit alpha, Phycocyanobilin lyase subunit beta | | Authors: | Hoeppner, A, Zhao, C, Xu, Q.-Z, Gaertner, W, Scheer, H, Zhao, K.-H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures and enzymatic mechanisms of phycobiliprotein lyases CpcE/F and PecE/F.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4YWC
 
 | | Crystal structure of Myc3(5-242) fragment in complex with Jaz9(218-239) peptide | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2015-03-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
5TZU
 
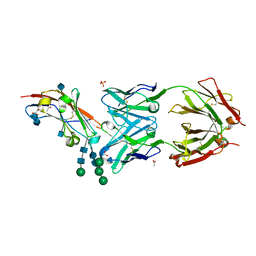 | | Crystal structure of human CD47 ECD bound to Fab of B6H12.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
3RHZ
 
 | |
5TZ2
 
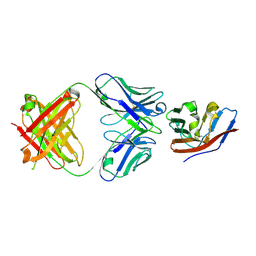 | | Crystal structure of human CD47 ECD bound to Fab of C47B222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C47B222 Fab Heavy Chain, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-21 | | Release date: | 2017-03-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
5TZT
 
 | | Crystal structure of human CD47 ECD bound to Fab of C47B161 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Heavy Chain of Fab C47B161, ... | | Authors: | Cardoso, R.M.F. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Anti-leukemic activity and tolerability of anti-human CD47 monoclonal antibodies.
Blood Cancer J, 7, 2017
|
|
7FHJ
 
 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
