3RZM
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 5'-D(*AP*TP*GP*TP*AP*TP*AP*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*TP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RZK
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*(EDA)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*TP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3RZJ
 
 | | Duplex Interrogation by a Direct DNA Repair Protein in the Search of Damage | | Descriptor: | 2-OXOGLUTARIC ACID, 5'-D(*CP*TP*GP*TP*CP*TP*(ME6)P*AP*CP*TP*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*AP*GP*TP*GP*AP*GP*AP*CP*A)-3', ... | | Authors: | Yi, C, Chen, B, Qi, B, Zhang, W, Jia, G, Zhang, L, Li, C, Dinner, A, Yang, C, He, C. | | Deposit date: | 2011-05-11 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Duplex interrogation by a direct DNA repair protein in search of base damage
Nat.Struct.Mol.Biol., 19, 2012
|
|
3NSS
 
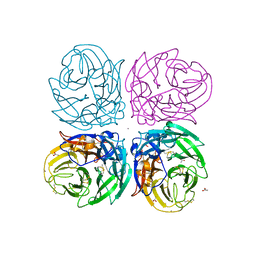 | | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active sites | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J.X, Zhang, W, Vavricka, C.J, Shi, Y, Gao, G.F. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KQ8
 
 | |
3ORR
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus | | Descriptor: | N5-carboxyaminoimidazole ribonucleotide synthetase | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
8HCZ
 
 | |
8HDF
 
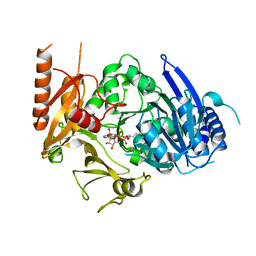 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8HD4
 
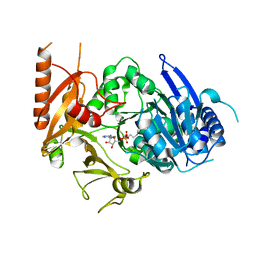 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
3ORS
 
 | | Crystal Structure of N5-Carboxyaminoimidazole Ribonucleotide Mutase from Staphylococcus aureus | | Descriptor: | N5-Carboxyaminoimidazole Ribonucleotide Mutase, SULFATE ION | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ORQ
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1N14
 
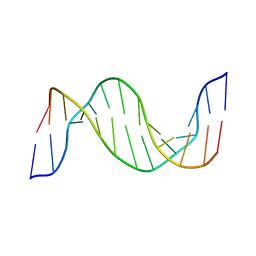 | | Structure and Dynamics of Thioguanine-modified Duplex DNA in Comparison with Unmodified DNA; Structure of Unmodified Duplex DNA | | Descriptor: | 5'-D(*GP*CP*TP*AP*AP*GP*GP*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | Authors: | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
3LTU
 
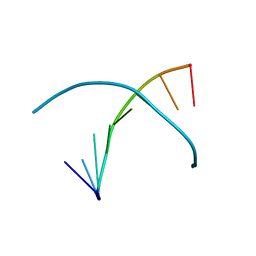 | | 5-SeMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5S)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
3LTR
 
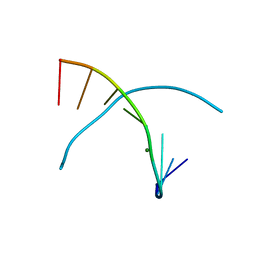 | | 5-OMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5O)P*AP*CP*AP*C)-3', MAGNESIUM ION | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
7N7X
 
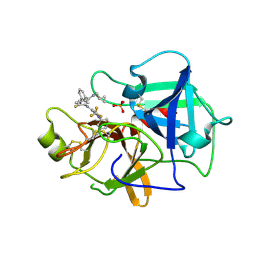 | | Crystal structure of BCX7353(ORLADEYO) in complex with human plasma kallikrein serine protease domain at 2.1 angstrom resolution | | Descriptor: | Orladeyo, PHOSPHATE ION, Plasma kallikrein light chain | | Authors: | Krishnan, R, Yarlagadda, B.S, Kotian, P, Polach, K.J, Zhang, W. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Berotralstat (BCX7353): Structure-Guided Design of a Potent, Selective, and Oral Plasma Kallikrein Inhibitor to Prevent Attacks of Hereditary Angioedema (HAE).
J.Med.Chem., 64, 2021
|
|
7M3Q
 
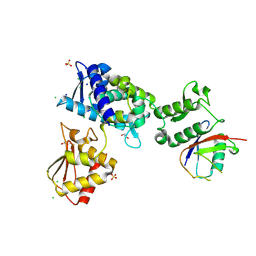 | | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV) | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Chowdhury, A, Singer, A.U, Ogunjimi, A.A, Teyra, J, Zhang, W, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-03-18 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Smurf2 HECT Domain with a High Affinity Ubiquitin Variant (UbV)
To be published
|
|
3K18
 
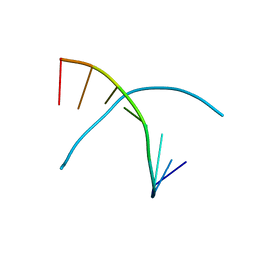 | | Tellurium modified DNA-8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(TTI)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Huang, Z. | | Deposit date: | 2009-09-25 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Synthesis and Structure Study of Oligonucleotides Derivatized with Tellurium Functionality
To be Published
|
|
2L89
 
 | |
1N17
 
 | | Structure and Dynamics of Thioguanine-modified Duplex DNA | | Descriptor: | 5'-D(*GP*CP*TP*AP*AP*GP*(S6G)P*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | Authors: | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
3O1R
 
 | | Iron-Catalyzed Oxidation Intermediates Captured in A DNA Repair Dioxygenase | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, DNA (5'-D(*AP*AP*CP*GP*GP*TP*AP*TP*TP*AP*CP*CP*T)-3'), ... | | Authors: | Yi, C, Jia, G, Hou, G, Dai, Q, Zhang, W, Zheng, G, Jian, X, Yang, C.-G, Cui, Q, He, C. | | Deposit date: | 2010-07-21 | | Release date: | 2010-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Iron-catalysed oxidation intermediates captured in a DNA repair dioxygenase.
Nature, 468, 2010
|
|
3FHQ
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Endo-beta-N-acetylglucosaminidase, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Jie, Y, Li, L, Shaw, N, Li, Y, Song, J, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.-C, Wang, L.-X, Wang, P, Liu, Z.-J. | | Deposit date: | 2008-12-10 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.452 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A
Plos One, 4, 2009
|
|
2LSP
 
 | | solution structures of BRD4 second bromodomain with NF-kB-K310ac peptide | | Descriptor: | Bromodomain-containing protein 4, NF-kB-K310ac peptide | | Authors: | Zhang, G, Liu, R, Zhong, Y, Plotnikov, A.N, Zhang, W, Rusinova, E, Gerona-Nevarro, G, Moshkina, N, Joshua, J, Chuang, P.Y, Ohlmeyer, M, He, J, Zhou, M.-M. | | Deposit date: | 2012-05-03 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Down-regulation of NF-kappa B transcriptional activity in HIV-associated kidney disease by BRD4 inhibition.
J.Biol.Chem., 287, 2012
|
|
5HTP
 
 | |
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
