2DHO
 
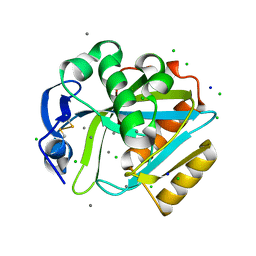 | | Crystal structure of human IPP isomerase I in space group P212121 | | Descriptor: | CHLORIDE ION, Isopentenyl-diphosphate delta-isomerase 1, MANGANESE (II) ION, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-03-24 | | Release date: | 2007-06-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
7E14
 
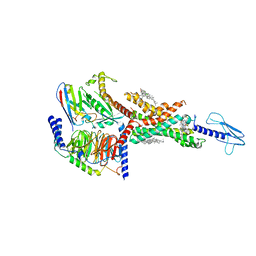 | | Compound2_GLP-1R_OWL833_Gs complex structure | | Descriptor: | 3-[(1S,2S)-1-(5-[(4S)-2,2-dimethyloxan-4-yl]-2-{(4S)-2-(4-fluoro-3,5-dimethylphenyl)-3-[3-(4-fluoro-1-methyl-1H-indazol-5-yl)-2-oxo-2,3-dihydro-1H-imidazol-1-yl]-4-methyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carbonyl}-1H-indol-1-yl)-2-methylcyclopropyl]-1,2,4-oxadiazol-5(4H)-one, CHOLESTEROL, G protein, ... | | Authors: | Cong, Z.T, Chen, L.N, Ma, H.L, Yang, D.H, Xu, H.E, Zhang, Y, Wang, M.W. | | Deposit date: | 2021-01-30 | | Release date: | 2021-07-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor.
Nat Commun, 12, 2021
|
|
7KQR
 
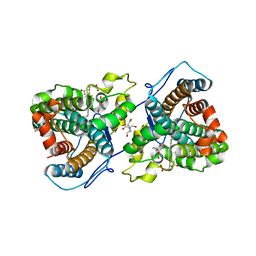 | | A 1.89-A resolution substrate-bound crystal structure of heme-dependent tyrosine hydroxylase from S. sclerotialus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heme-dependent L-tyrosine hydroxylase, ... | | Authors: | Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQU
 
 | | A 1.58-A resolution crystal structure of ferric-hydroperoxo intermediate of L-tyrosine hydroxylase in complex with 3-fluoro-L-tyrosine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Davis, I, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
7KQT
 
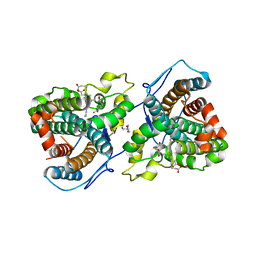 | |
7KQS
 
 | | A 1.68-A resolution 3-fluoro-L-tyrosine bound crystal structure of heme-dependent tyrosine hydroxylase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Wang, Y, Liu, A. | | Deposit date: | 2020-11-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.677 Å) | | Cite: | Molecular Rationale for Partitioning between C-H and C-F Bond Activation in Heme-Dependent Tyrosine Hydroxylase.
J.Am.Chem.Soc., 143, 2021
|
|
2ZMV
 
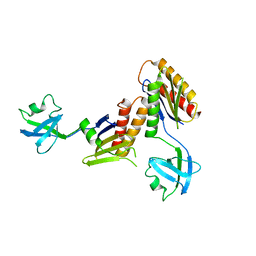 | | Crystal structure of Synbindin | | Descriptor: | Trafficking protein particle complex subunit 4 | | Authors: | Fan, F. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human synbindin reveals two conformations of longin domain
Biochem.Biophys.Res.Commun., 378, 2009
|
|
7KID
 
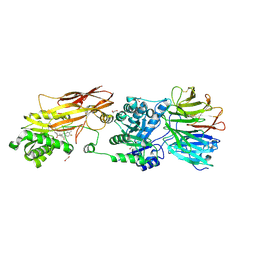 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 72 | | Descriptor: | (1S,2R,3aR,4S,6aR)-4-[(2-amino-3,5-difluoroquinolin-7-yl)methyl]-2-(4-amino-5-fluoro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydropentalene-1,6a(1H)-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7KIC
 
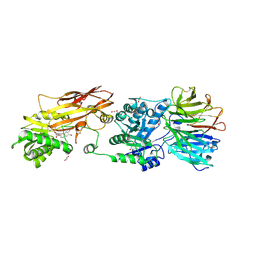 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 34 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-methyl-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
7KIB
 
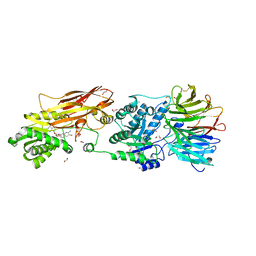 | | PRMT5:MEP50 Complexed with 5,5-Bicyclic Inhibitor Compound 4 | | Descriptor: | (2R,3R,3aS,6S,6aR)-6-[(2-amino-3-bromoquinolin-7-yl)oxy]-2-(4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)hexahydro-3aH-cyclopenta[b]furan-3,3a-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Palte, R.L, Hayes, R.P. | | Deposit date: | 2020-10-23 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Discovery of Two Novel Classes of 5,5-Bicyclic Nucleoside-Derived PRMT5 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
2FEK
 
 | | Structure of a protein tyrosine phosphatase | | Descriptor: | Low molecular weight protein-tyrosine-phosphatase wzb | | Authors: | Lescop, E, Jin, C. | | Deposit date: | 2005-12-16 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Escherichia coli Wzb reveals a novel substrate recognition mechanism of prokaryotic low molecular weight protein-tyrosine phosphatases
J.Biol.Chem., 281, 2006
|
|
3BLI
 
 | |
6CBV
 
 | | Crystal structure of BRIL bound to an affinity matured synthetic antibody. | | Descriptor: | BRIL, FORMIC ACID, GLYCEROL, ... | | Authors: | Mukherjee, S, Skrobek, B, Kossiakoff, A.A. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
6JQ1
 
 | | Crystal Structure of DdrO from Deinococcus geothermalis | | Descriptor: | LITHIUM ION, Transcriptional regulator, XRE family | | Authors: | Lu, H, Hua, Y, Zhao, Y. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO.
Nucleic Acids Res., 47, 2019
|
|
4HRV
 
 | |
5ZYQ
 
 | | The Structure of Human PAF1/CTR9 complex | | Descriptor: | RNA polymerase-associated protein CTR9 homolog,RNA polymerase II-associated factor 1 homolog | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.531 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
5ZYP
 
 | | Structure of the Yeast Ctr9/Paf1 complex | | Descriptor: | NICKEL (II) ION, RNA polymerase-associated protein CTR9,RNA polymerase II-associated protein 1 | | Authors: | Xie, Y, Zheng, M, Zhou, H, Long, J. | | Deposit date: | 2018-05-28 | | Release date: | 2018-09-26 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation.
Nat Commun, 9, 2018
|
|
7CZ5
 
 | | Cryo-EM structure of the human growth hormone-releasing hormone receptor-Gs protein complex | | Descriptor: | CHOLESTEROL, Growth hormone-releasing hormone receptor,growth hormone-releasing hormone receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhou, F, Zhang, H, Cong, Z, Zhao, L, Zhou, Q, Mao, C, Cheng, X, Shen, D, Cai, X, Ma, C, Wang, Y, Dai, A, Zhou, Y, Sun, W, Zhao, F, Zhao, S, Jiang, H, Jiang, Y, Yang, D, Xu, H.E, Zhang, Y, Wang, M. | | Deposit date: | 2020-09-07 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for activation of the growth hormone-releasing hormone receptor.
Nat Commun, 11, 2020
|
|
8I6T
 
 | | The cryo-EM structure of OsCyc1 hexamer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8I6U
 
 | | The cryo-EM structure of OsCyc1 dimer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8IH5
 
 | |
8I6P
 
 | | The cryo-EM structure of OsCyc1 tetramer state | | Descriptor: | Syn-copalyl diphosphate synthase, chloroplastic | | Authors: | Ma, X.L, Xu, H.F, Tong, Y.R, Luo, Y.F, Dong, Q.H, Jiang, T. | | Deposit date: | 2023-01-29 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional investigations of syn-copalyl diphosphate synthase from Oryza sativa.
Commun Chem, 6, 2023
|
|
8KBW
 
 | |
3F6G
 
 | | Crystal structure of the regulatory domain of LiCMS in complexed with isoleucine - type II | | Descriptor: | Alpha-isopropylmalate synthase, ISOLEUCINE, SULFATE ION, ... | | Authors: | Zhang, P, Ma, J, Zhao, G, Ding, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of the inhibitor selectivity and insights into the feedback inhibition mechanism of citramalate synthase from Leptospira interrogans
Biochem.J., 421, 2009
|
|
2X06
 
 | | SULFOLACTATE DEHYDROGENASE FROM METHANOCALDOCOCCUS JANNASCHII | | Descriptor: | L-SULFOLACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D. | | Deposit date: | 2009-12-07 | | Release date: | 2009-12-15 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Methanoarchaeal Sulfolactate Dehydrogenase: Prototype of a New Family of Nadh-Dependent Enzymes.
Embo J., 23, 2004
|
|
