8XVI
 
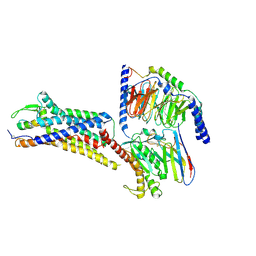 | | Cryo-EM structure of ETAR bound with Endothelin1 | | Descriptor: | Endoglucanase H,Endothelin-1 receptor, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVH
 
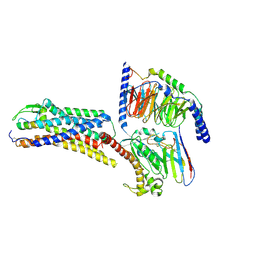 | | Cryo-EM structure of ETBR bound with Endothelin1 | | Descriptor: | Endothelin-1, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8XVE
 
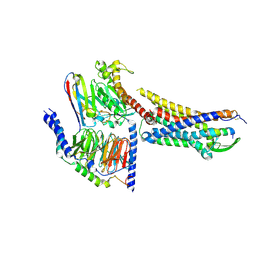 | | Cryo-EM structure of ETBR bound with BQ3020 | | Descriptor: | BQ3020, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
8UV0
 
 | |
8HJ2
 
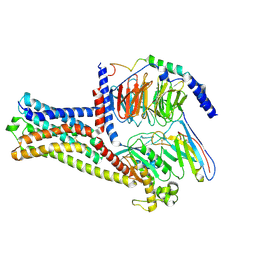 | | GPR21 wt with G15 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HIX
 
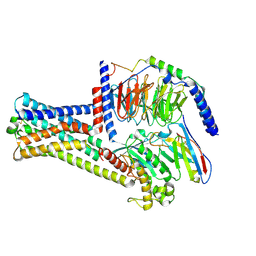 | | Cryo-EM structure of GPR21_m5_Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X, Xu, F. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8HJ0
 
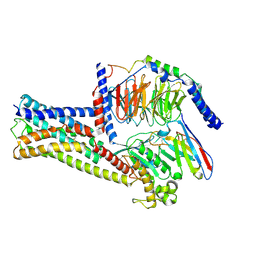 | | GPR21(m5) and G15 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Chen, B, Lin, X. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structures of orphan GPR21 signaling complexes.
Nat Commun, 14, 2023
|
|
8WRZ
 
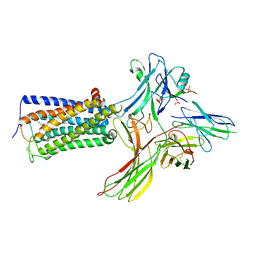 | | Cry-EM structure of cannabinoid receptor-beta-arrestin-1 complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Beta-arrestin-1, Soluble cytochrome b562,Cannabinoid receptor 1, ... | | Authors: | Wang, Y.X, Wang, T, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of cannabinoid receptor CB1-beta-arrestin complex.
Protein Cell, 15, 2024
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
4DMN
 
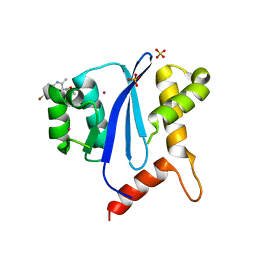 | | HIV-1 Integrase Catalytical Core Domain | | Descriptor: | (2S)-[6-bromo-4-(4-chlorophenyl)-2-methylquinolin-3-yl](methoxy)ethanoic acid, ARSENIC, HIV-1 Integrase, ... | | Authors: | Feng, L, Kvaratskhelia, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors.
J.Biol.Chem., 287, 2012
|
|
6KFW
 
 | |
6TJK
 
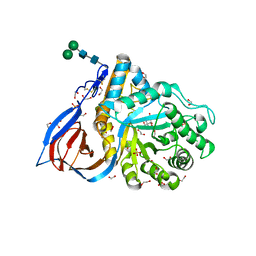 | | Crystal Structure of Recombinant GBA in Complex with Bis-Tris Propane. | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-11-26 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6LGT
 
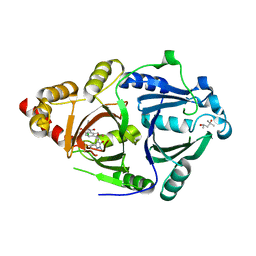 | | Complex structure of HPPD with an inhibitor Y16542 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-(2-chlorophenyl)-6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-1,5-dimethyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.Y, Yang, W.C, Yang, G.F. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Discovery of Novel Pyrazole-Quinazoline-2,4-dione Hybrids as 4-Hydroxyphenylpyruvate Dioxygenase Inhibitors.
J.Agric.Food Chem., 68, 2020
|
|
4RDD
 
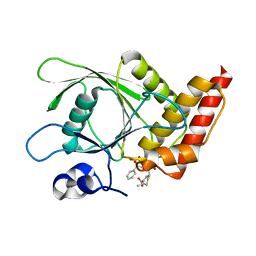 | | Co-crystal structure of SHP2 in complex with a Cefsulodin derivative | | Descriptor: | 1-({(2R)-4-carboxy-2-[(R)-carboxy{[(2R)-2-phenyl-2-sulfoacetyl]amino}methyl]-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.Y, Yu, Z.H, He, R, Zhang, R.Y. | | Deposit date: | 2014-09-18 | | Release date: | 2015-07-01 | | Last modified: | 2015-12-16 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Exploring the Existing Drug Space for Novel pTyr Mimetic and SHP2 Inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
7XX4
 
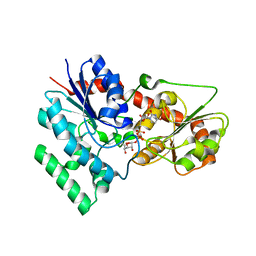 | | designed glycosyltransferase | | Descriptor: | Oleandomycin glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Lu, M, Wu, X. | | Deposit date: | 2022-05-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Design of a chimeric glycosyltransferase OleD for the site-specific O-monoglycosylation of 3-hydroxypyridine in nosiheptide.
Microb Biotechnol, 16, 2023
|
|
7E5X
 
 | |
4J51
 
 | |
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp4/5 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp14/15 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp15/16 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp9/10 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVX
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp6|7 peptidyl substrate | | Descriptor: | 3C-like proteinase, nsp6/7 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVW
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp5|6 peptidyl substrate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, nsp5/6 peptidyl substrate | | Authors: | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | Deposit date: | 2021-01-15 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6M1V
 
 | |
