8IJR
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with diacylglycerol/phosphoethanolamine | | Descriptor: | (2S)-1-(hexadecanoyloxy)-3-hydroxypropan-2-yl (11Z)-octadec-11-enoate, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IJQ
 
 | | The cryo-EM structure of human sphingomyelin synthase-related protein in complex with ceramide | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, Sphingomyelin synthase-related protein 1 | | Authors: | Hu, K, Zhang, Q, Chen, Y, Yao, D, Zhou, L, Cao, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM structure of human sphingomyelin synthase and its mechanistic implications for sphingomyelin synthesis.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6A96
 
 | | Cryo-EM structure of the human alpha5beta3 GABAA receptor in complex with GABA and Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-5,Gamma-aminobutyric acid receptor subunit alpha-5, ... | | Authors: | Liu, S, Xu, L, Guan, F, Liu, Y.T, Cui, Y, Zhang, Q, Bi, G.Q, Zhou, Z.H, Zhang, X, Ye, S. | | Deposit date: | 2018-07-11 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM structure of the human alpha 5 beta 3 GABAAreceptor.
Cell Res., 28, 2018
|
|
3TK3
 
 | | Cytochrome P450 2B4 mutant L437A in complex with 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gay, S.C, Jang, H.H, Wilderman, P.R, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-08-25 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme.
Febs J., 279, 2012
|
|
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HWL
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5Z2V
 
 | | Crystal structure of RecR from Pseudomonas aeruginosa PAO1 | | Descriptor: | ANY 5'-MONOPHOSPHATE NUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Che, S, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of RecR, a member of the RecFOR DNA-repair pathway, from Pseudomonas aeruginosa PAO1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7C81
 
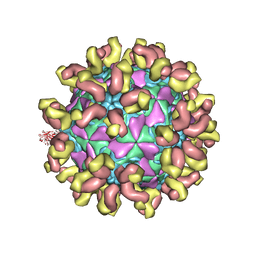 | | E30 F-particle in complex with 6C5 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
6INB
 
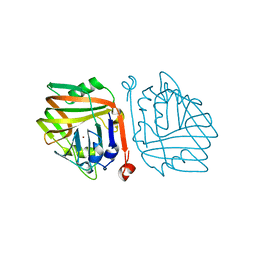 | |
6INC
 
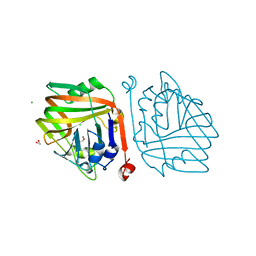 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
7VH8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Zhao, Y, Zhang, Q, Yang, H, Rao, Z. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332.
Protein Cell, 13, 2022
|
|
3SZ5
 
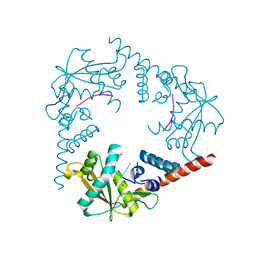 | | Crystal Structure of LHK-Exo in complex with 5-phosphorylated oligothymidine (dT)4 | | Descriptor: | 5'-D(P*TP*TP*TP*T)-3', Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
7V5H
 
 | |
6JC4
 
 | |
3SYY
 
 | | Crystal Structure of an alkaline exonuclease (LHK-Exo) from Laribacter hongkongensis | | Descriptor: | Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3SZ4
 
 | | Crystal Structure of LHK-Exo in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
7V9Z
 
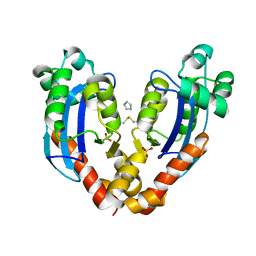 | |
7VA3
 
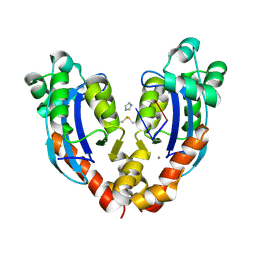 | |
7VA6
 
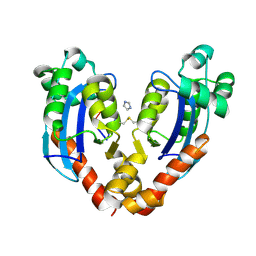 | |
7VA2
 
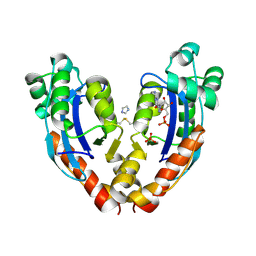 | |
7C80
 
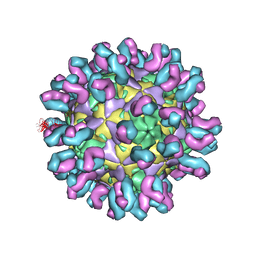 | | E30 F-particle in complex with 4B10 | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Wang, K, Zheng, B, Zhang, L, Cui, L, Su, X, Zhang, Q, Guo, Y, Zhu, L, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.
Nat Commun, 11, 2020
|
|
7X78
 
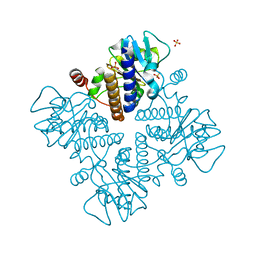 | | L-fuculose 1-phosphate aldolase | | Descriptor: | L-fuculose phosphate aldolase, MAGNESIUM ION, SULFATE ION | | Authors: | Lou, X, Zhang, Q, Bartlam, M. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of an L-fuculose-1-phosphate aldolase from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 607, 2022
|
|
2KKS
 
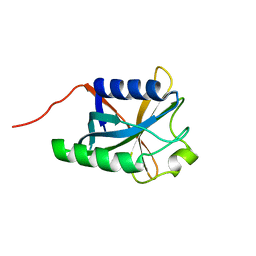 | | Solution Structure Of Protein DSY2949 From Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR27 | | Descriptor: | Uncharacterized protein | | Authors: | Wu, Y, Mills, J.L, Wang, H, Ciccosanti, C, Jiang, M, Sukumaran, D, Zhang, Q, Nair, R, Rost, B, Acton, T, Xiao, R, Swapna, G.V.T, Everett, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein DSY2949 From Desulfitobacterium hafniense. Northeast Structural Genomics Consortium Target DhR27
To be Published
|
|
2K95
 
 | | Solution structure of the wild-type P2B-P3 pseudoknot of human telomerase RNA | | Descriptor: | Telomerase RNA P2b-P3 pseudoknot | | Authors: | Kim, N.-K, Zhang, Q, Zhou, J, Theimer, C.A, Peterson, R.D, Feigon, J. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Wild-type Pseudoknot of Human Telomerase RNA.
J.Mol.Biol., 384, 2008
|
|
2KHN
 
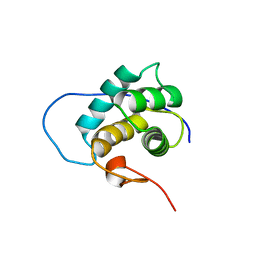 | | NMR solution structure of the EH 1 domain from human intersectin-1 protein. Northeast Structural Genomics Consortium target HR3646E. | | Descriptor: | Intersectin-1 | | Authors: | Mills, J.L, Ghosh, A, Garcia, E, Zhang, Q, Shastry, R, Foote, E.L, Janjua, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-04-09 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the EH 1 domain from human intersectin-1 protein. Northeast Structural Genomics Consortium target HR3646E.
To be Published
|
|
