3FM5
 
 | | X-ray crystal structure of transcriptional regulator (MarR family) from Rhodococcus sp. RHA1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nocek, B, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of transcriptional regulator (MarR family) from Rhodococcus sp. RHA1
To be Published
|
|
5L0L
 
 | | Crystal structure of Uncharacterized protein LPG0439 | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | Authors: | Chang, C, Skarina, T, Khutoreskaya, G, Savchenko, A, Joachimiak, A. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Uncharacterized protein LPG0439
To Be Published
|
|
3NKD
 
 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | CRISPR-associated protein Cas1 | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
3FYB
 
 | | Crystal structure of a protein of unknown function (DUF1244) from Alcanivorax borkumensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a protein of unknown function (DUF1244) from Alcanivorax borkumensis
To be Published
|
|
3FZV
 
 | | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa | | Descriptor: | Probable transcriptional regulator, SULFATE ION | | Authors: | Knapik, A.A, Tkaczuk, K.L, Chruszcz, M, Wang, S, Zimmerman, M.D, Cymborowski, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Bujnicki, J.M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of PA01 protein, putative LysR family transcriptional regulator from Pseudomonas aeruginosa
To be Published
|
|
3NI7
 
 | | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718 | | Descriptor: | Bacterial regulatory proteins, TetR family | | Authors: | Knapik, A, Chruszcz, M, Cymborowski, M, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718
To be Published
|
|
3NKE
 
 | | High resolution structure of the C-terminal domain CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, SULFITE ION, ... | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
2G8Y
 
 | | The structure of a putative malate/lactate dehydrogenase from E. coli. | | Descriptor: | 1,2-ETHANEDIOL, Malate/L-lactate dehydrogenases, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of a putative malate/lactate dehydrogenase from E. coli.
To be Published
|
|
1XSV
 
 | | X-ray crystal structure of conserved hypothetical UPF0122 protein SAV1236 from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | Hypothetical UPF0122 protein SAV1236 | | Authors: | Walker, J.R, Xu, X, Virag, C, McDonald, M.-L, Houston, S, Buzadzija, K, Vedadi, M, Dharamsi, A, Fiebig, K.M, Savchenko, A. | | Deposit date: | 2004-10-20 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Crystal Structure of Conserved Hypothetical UPF0122 Protein SAV1236 From Staphylococcus aureus
To be Published
|
|
1TE2
 
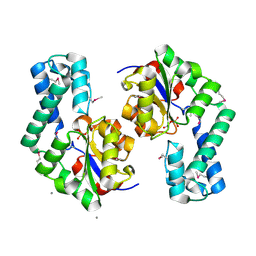 | | Putative Phosphatase Ynic from Escherichia coli K12 | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 2-deoxyglucose-6-P phosphatase, CALCIUM ION | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Putative Phosphatase Ynic from Escherichia coli K12
To be Published
|
|
3BRM
 
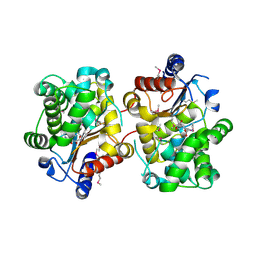 | | Crystal structure of the covalent complex between the Bacillus subtilis glutaminase YbgJ and 5-oxo-L-norleucine formed by reaction of the protein with 6-diazo-5-oxo-L-norleucine | | Descriptor: | 5-OXO-L-NORLEUCINE, Glutaminase 1 | | Authors: | Singer, A.U, Kim, Y, Dementieva, I, Vinokour, E, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-05-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
5CYV
 
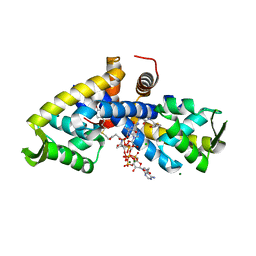 | | Crystal structure of CouR from Rhodococcus jostii RHA1 bound to p-coumaroyl-CoA | | Descriptor: | ACETATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The activity of CouR, a MarR family transcriptional regulator, is modulated through a novel molecular mechanism.
Nucleic Acids Res., 44, 2016
|
|
4U12
 
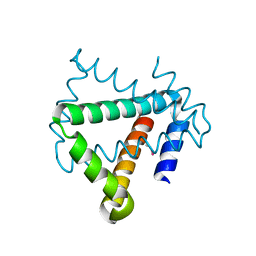 | | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer | | Descriptor: | Uncharacterized protein HP0242 | | Authors: | Grabowski, M, Shabalin, I.G, Chruszcz, M, Skarina, T, Onopriyenko, O, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-14 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer
to be published
|
|
2AF7
 
 | | Crystal structure of the gamma-carboxymuconolactone decarboxylase from Methanobacterium thermoautotrophicum. Northeast Structural Genomics Consortium target TT747. | | Descriptor: | gamma-carboxymuconolactone decarboxylase | | Authors: | Vorobiev, S.M, Kuzin, A, Skarina, T, Savchenko, A, Semesi, A, Arrowsmith, C, Edwards, A, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-07-25 | | Release date: | 2005-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal structure of the gamma-carboxymuconolactone decarboxylase from Methanobacterium thermoautotrophicum.
To be Published
|
|
3D1R
 
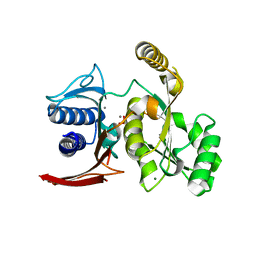 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
2LF6
 
 | | Solution NMR structure of HopABPph1448_220_320 from Pseudomonas syringae pv. phaseolicola str. 1448A, Midwest Center for Structural Genomics target APC40132.4 and Northeast Structural Genomics Consortium target PsT3A | | Descriptor: | Effector protein hopAB1 | | Authors: | Wu, B, Yee, A, Houliston, S, Semesi, A, Garcia, M, Singer, A.U, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2011-06-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
1Q7H
 
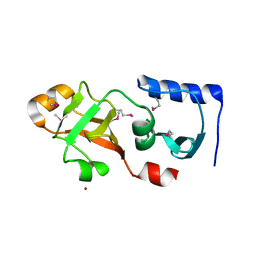 | | Structure of a Conserved PUA Domain Protein from Thermoplasma acidophilum | | Descriptor: | ZINC ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-18 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a conserved hypothetical protein from T. acidophilum
TO BE PUBLISHED
|
|
1ZL0
 
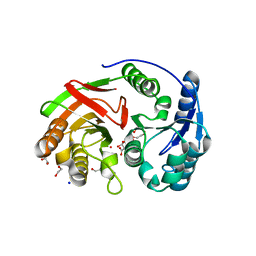 | | Structure of Protein of Unknown Function PA5198 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-04 | | Release date: | 2005-06-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystal structure of hypothetical protein PA5198 at 1.1 A resolution.
To be Published
|
|
6P2K
 
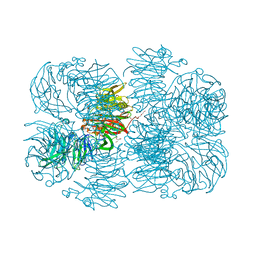 | | Crystal structure of AFV00434, an ancestral GH74 enzyme | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
4Q3K
 
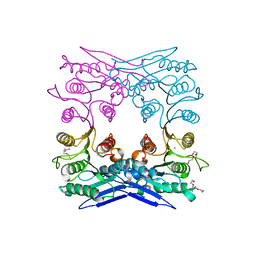 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
6P2O
 
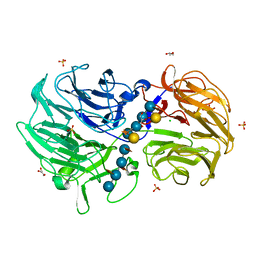 | | Crystal structure of Streptomyces rapamycinicus GH74 in complex with xyloglucan fragments XLLG and XXXG | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
4Q3L
 
 | | Crystal structure of MGS-M2, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | GLYCEROL, MGS-M2 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
2A61
 
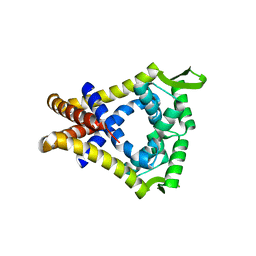 | | The crystal structure of transcriptional regulator Tm0710 from Thermotoga maritima | | Descriptor: | transcriptional regulator Tm0710 | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Chang, C, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-01 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of transcriptional regulator Tm0710 from Thermotoga maritima
To be Published
|
|
2P06
 
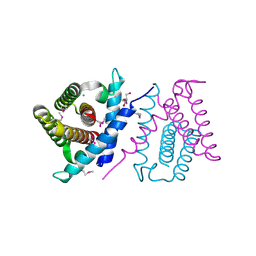 | | Crystal structure of a predicted coding region AF_0060 from Archaeoglobus fulgidus DSM 4304 | | Descriptor: | GLYCEROL, Hypothetical protein AF_0060, MAGNESIUM ION | | Authors: | Nocek, B, Xu, X, Koniyenko, Y, Yakounine, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-28 | | Release date: | 2007-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a predicted coding region AF_0060 from Archaeoglobus fulgidus DSM 4304
To be Published
|
|
3KYE
 
 | | Crystal Structure of Roadblock/LC7 Domain from Streptomyces avermitilis | | Descriptor: | Roadblock/LC7 domain, Robl_LC7 | | Authors: | Kim, Y, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-05 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Roadblock/LC7 Domain from Streptomyces avermitilis
To be Published
|
|
