7ALG
 
 | | The RSLex - sulfonato-calix[8]arene complex, P3 form, acetate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
7ALF
 
 | | The dimethylated RSL - sulfonato-calix[8]arene complex, P3 form, acetate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2020-10-06 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.261 Å) | | Cite: | Facile Fabrication of Protein-Macrocycle Frameworks.
J.Am.Chem.Soc., 143, 2021
|
|
6ZUL
 
 | | Crystal structure of dimethylated RSL in complex with cucurbit[7]uril and zinc | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
6ZUM
 
 | | Crystal structure of dimethylated RSL-N23H (RSL-B3) in complex with cucurbit[7]uril and zinc | | Descriptor: | ACETATE ION, Fucose-binding lectin protein, SODIUM ION, ... | | Authors: | Guagnini, F, Engilberge, S, Flood, R.J, Ramberg, K.O, Crowley, P.B. | | Deposit date: | 2020-07-23 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Metal-Mediated Protein-Cucurbituril Crystalline Architectures
Cryst.Growth Des., 2020
|
|
6STZ
 
 | |
6SU0
 
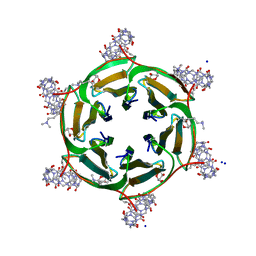 | | Crystal structure of dimethylated RSLex in complex with cucurbit[7]uril | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, cucurbit[7]uril, ... | | Authors: | Guagnini, F, Engilberge, S, Crowley, P.B. | | Deposit date: | 2019-09-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineered assembly of a protein-cucurbituril biohybrid.
Chem.Commun.(Camb.), 56, 2020
|
|
6Q3T
 
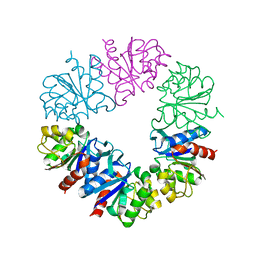 | | Structure of Protease1 from Pyrococcus horikoshii at room temperature in ChipX microfluidic device | | Descriptor: | Deglycase PH1704 | | Authors: | de Wijn, R, Engilberge, S, Olieric, V, Girard, E, Sauter, C. | | Deposit date: | 2018-12-04 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
6GSB
 
 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
6GSC
 
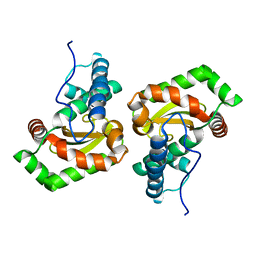 | | Sphingobacterium sp. T2 manganese superoxide dismutase catalyses the oxidative demethylation of polymeric lignin via generation of hydroxyl radical | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Rashid, G.M.M, Zhang, X, Wilkinson, R.C, Fulop, V, Cottyn, B, Baumberger, S, Bugg, T.D.H. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Sphingobacterium sp. T2 Manganese Superoxide Dismutase Catalyzes the Oxidative Demethylation of Polymeric Lignin via Generation of Hydroxyl Radical.
ACS Chem. Biol., 13, 2018
|
|
6S8Y
 
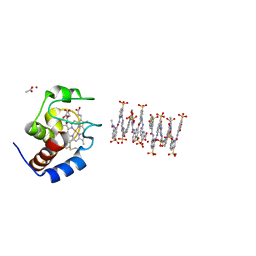 | | Crystal structure of cytochrome c in complex with a sulfonated quinoline-derived foldamer | | Descriptor: | 8-acetamido-2-[[2-[[2-[[2-[[2-[[2-[[2-[(2-carboxy-4-sulfonato-quinolin-8-yl)carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]-4-sulfonato-quinolin-8-yl]carbamoyl]quinoline-4-sulfonate, ACETATE ION, Cytochrome c iso-1, ... | | Authors: | Alex, J.M, Corvaglia, V, Hu, X, Engilberge, S, Huc, I, Crowley, P.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of a protein-aromatic foldamer composite: macromolecular chiral resolution.
Chem.Commun.(Camb.), 55, 2019
|
|
6STH
 
 | |
7B1S
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum at 0.994-A resolution | | Descriptor: | (2S)-2-{[(2S)-2-{[(2S)-2-hydroxypropyl]oxy}propyl]oxy}propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-25 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.992 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
7B2C
 
 | | Crystal structure of the ethyl-coenzyme M reductase from Candidatus Ethanoperedens thermophilum gassed with xenon | | Descriptor: | (2R)-2-[(2S)-2-[(2S)-2-oxidanylpropoxy]propoxy]propan-1-ol, 1-THIOETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-26 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
7B2H
 
 | | Crystal structure of the methyl-coenzyme M reductase from Methanothermobacter Marburgensis derivatized with xenon | | Descriptor: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Wagner, T, Lemaire, O.N, Engilberge, S. | | Deposit date: | 2020-11-27 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure of a key enzyme for anaerobic ethane activation.
Science, 373, 2021
|
|
7AZ5
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZG
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 4 bound | | Descriptor: | Beta sliding clamp, Peptide 4 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZL
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 38 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ7
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | Descriptor: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | Descriptor: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ8
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 43 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZD
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 20 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZE
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 18 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, MALONATE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZC
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 22 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, Peptide 22 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZF
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 8 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZK
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 35 bound | | Descriptor: | Beta sliding clamp, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
