3QTF
 
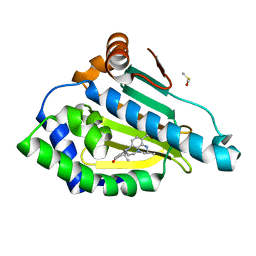 | | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity | | Descriptor: | (6S)-6,15,15,18-tetramethyl-17-oxo-2,3,4,5,6,7,14,15,16,17-decahydro-1H-8,12-(metheno)[1,4,9]triazacyclotetradecino[9,8-a]indole-9-carboxamide, DIMETHYL SULFOXIDE, Heat shock protein HSP 90-alpha | | Authors: | Zapf, C.W, Bloom, J.D, McBean, J.L, Dushin, R.G, Nittoli, T, Ingalls, C, Sutherland, A.G, Sonye, J.P, Eid, C.N, Golas, J, Liu, H, Boschelli, F, Hu, Y, Vogan, E.M, Levin, J.I. | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5703 Å) | | Cite: | Design and SAR of macrocyclic Hsp90 inhibitors with increased metabolic stability and potent cell-proliferation activity.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3TEK
 
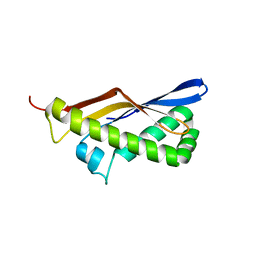 | | ThermoDBP: a non-canonical single-stranded DNA binding protein with a novel structure and mechanism | | Descriptor: | ThermoDBP-single stranded DNA binding protein | | Authors: | White, M.F, Paytubi, S, Liu, H, Graham, S, McMahon, S.A, Naismith, J.H. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MLB
 
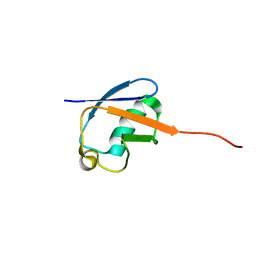 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | Descriptor: | redesigned ubiquitin | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MN4
 
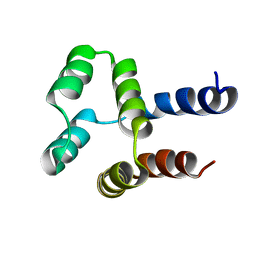 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | Descriptor: | Computational designed protein based on structure template 1cy5 | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
3VPN
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
3VPM
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
2IX2
 
 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2JG2
 
 | | HIGH RESOLUTION STRUCTURE OF SPT WITH PLP INTERNAL ALDIMINE | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-05-01 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
4Q5W
 
 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
3ZIF
 
 | | Cryo-EM structures of two intermediates provide insight into adenovirus assembly and disassembly | | Descriptor: | HEXON PROTEIN, PENTON PROTEIN, PIX, ... | | Authors: | Cheng, L, Huang, X, Li, X, Xiong, W, Sun, W, Yang, C, Zhang, K, Wang, Y, Liu, H, Ji, G, Sun, F, Zheng, C, Zhu, P. | | Deposit date: | 2013-01-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-Em Structures of Two Bovine Adenovirus Type 3 Intermediates
Virology, 450, 2014
|
|
2JGT
 
 | | Low resolution structure of SPT | | Descriptor: | SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-14 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
5ZTL
 
 | | Non-cryogenic structure of light-driven chloride pump having an NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Park, S.Y, Liu, H, Lee, W. | | Deposit date: | 2018-05-04 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Non-cryogenic structure of a chloride pump provides crucial clues to temperature-dependent channel transport efficiency
J. Biol. Chem., 294, 2019
|
|
3ZFV
 
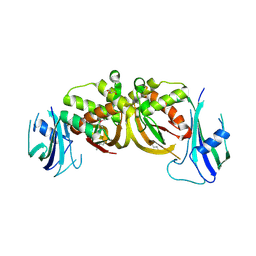 | | Crystal structure of an archaeal CRISPR-associated Cas6 nuclease | | Descriptor: | CRISPR-ASSOCIATED ENDORIBONUCLEASE CAS6 1, GLYCEROL | | Authors: | Reeks, J, Liu, H, White, M.F, Naismith, J.H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Biochem.J., 452, 2013
|
|
4A3O
 
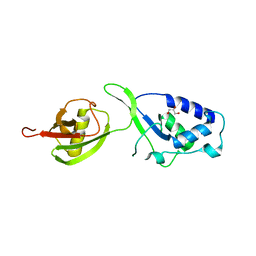 | | Crystal structure of the USP15 DUSP-UBL monomer | | Descriptor: | GLYCEROL, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
4A3P
 
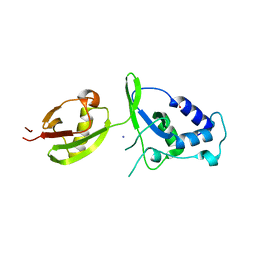 | | Structure of USP15 DUSP-UBL deletion mutant | | Descriptor: | ACETATE ION, IODIDE ION, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
4AVR
 
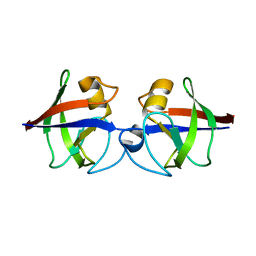 | | Crystal structure of the hypothetical protein Pa4485 from Pseudomonas aeruginosa | | Descriptor: | PA4485 | | Authors: | McMahon, S.A, Moynie, L, Liu, H, Duthie, F, Alphey, M.S, Naismith, J.H. | | Deposit date: | 2012-05-29 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | The Aeropath Project Targeting Pseudomonas Aeruginosa: Crystallographic Studies for Assessment of Potential Targets in Early-Stage Drug Discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6JLE
 
 | | Crystal structure of MORN4/Myo3a complex | | Descriptor: | CITRIC ACID, GLYCEROL, MORN repeat-containing protein 4, ... | | Authors: | Li, J, Liu, H, Raval, M.H, Wan, J, Yengo, C.M, Liu, W, Zhang, M. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the MORN4/Myo3a Tail Complex Reveals MORN Repeats as Protein Binding Modules.
Structure, 27, 2019
|
|
8HND
 
 | | Cryo-EM structure of human OATP1B1 in complex with estrone-3-sulfate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier organic anion transporter family member 1B1, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8HNC
 
 | | Cryo-EM structure of human OATP1B1 in complex with bilirubin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Shan, Z, Yang, X, Liu, H, Nan, J, Yuan, Y, Zhang, Y. | | Deposit date: | 2022-12-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Cryo-EM structures of human organic anion transporting polypeptide OATP1B1.
Cell Res., 33, 2023
|
|
8JPC
 
 | | cryo-EM structure of NTSR1-GRK2-Galpha(q) complexes 2 | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-adrenergic receptor kinase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
8JPD
 
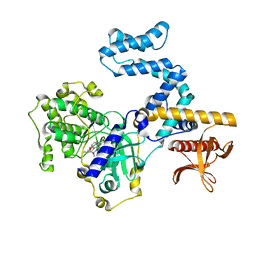 | | Focused refinement structure of GRK2 in NTSR1-GRK2-Galpha(q) complexes | | Descriptor: | Beta-adrenergic receptor kinase 1, STAUROSPORINE | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
8JPB
 
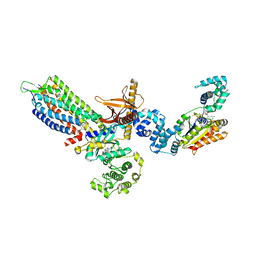 | | cryo-EM structure of NTSR1-GRK2-Galpha(q) complexes 1 | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, Beta-adrenergic receptor kinase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
8JPE
 
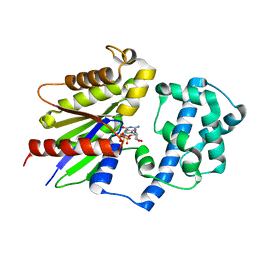 | | Focused refinement structure of Galpha(q) in NTSR1-GRK2-Galpha(q) complexes | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(q) subunit alpha, MAGNESIUM ION, ... | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
8JPF
 
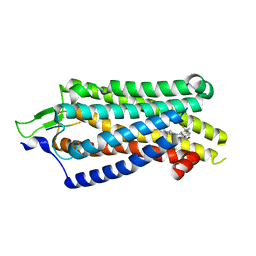 | | Focused refiment structure of NTSR1 in NTSR1-GRK2-Galpha(q) complexes | | Descriptor: | 2-[{2-(1-fluorocyclopropyl)-4-[4-(2-methoxyphenyl)piperidin-1-yl]quinazolin-6-yl}(methyl)amino]ethan-1-ol, NTS, Neurotensin receptor type 1 | | Authors: | Duan, J, Liu, H, Zhao, F, Yuan, Q, Ji, Y, Xu, H.E. | | Deposit date: | 2023-06-11 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | GPCR activation and GRK2 assembly by a biased intracellular agonist.
Nature, 620, 2023
|
|
