7EA4
 
 | |
2Y04
 
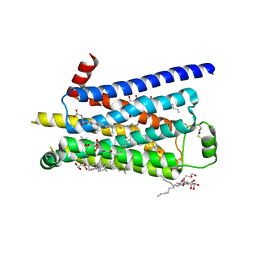 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND PARTIAL AGONIST SALBUTAMOL | | Descriptor: | BETA-1 ADRENERGIC RECEPTOR, CHOLESTEROL HEMISUCCINATE, HEGA-10, ... | | Authors: | Warne, A, Moukhametzianov, R, Baker, J.G, Nehme, R, Edwards, P.C, Leslie, A.G.W, Schertler, G.F.X, Tate, C.G. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Structural Basis for Agonist and Partial Agonist Action on a Beta1-Adrenergic Receptor
Nature, 469, 2011
|
|
5XXO
 
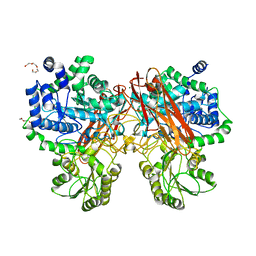 | | Crystal structure of mutant (D286N) GH3 beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorotriose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahash, Y, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
4JB9
 
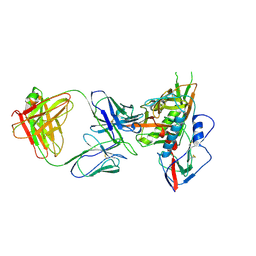 | | Crystal structure of antibody VRC06 in complex with HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, antibody VRC06 heavy chain, antibody VRC06 light chain, ... | | Authors: | Kwon, Y.D, Zhou, T, Srivatsan, S, Kwong, P.D. | | Deposit date: | 2013-02-19 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization.
Science, 340, 2013
|
|
4J6R
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC23 in complex with HIV-1 gp120 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-02-11 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization.
Science, 340, 2013
|
|
5GXW
 
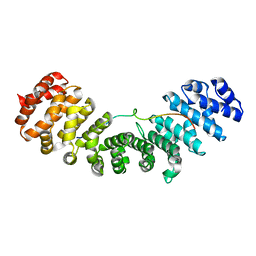 | | Importin and NuMA complex | | Descriptor: | Importin subunit alpha-1, Peptide from Nuclear mitotic apparatus protein 1 | | Authors: | Chang, C.-C, Huang, T.-L, Hsia, K.-C. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Regulation of mitotic spindle assembly factor NuMA by Importin-beta
J. Cell Biol., 216, 2017
|
|
5ELO
 
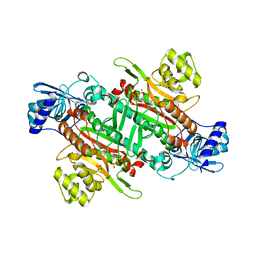 | |
5ELN
 
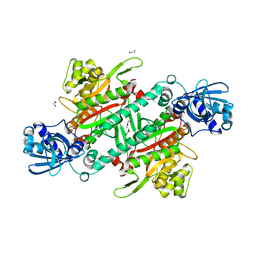 | |
7YFC
 
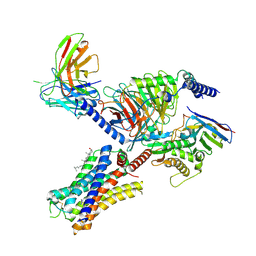 | | Cryo-EM structure of the histamine-bound histamine H4 receptor and Gq complex | | Descriptor: | CHOLESTEROL, Engineered G-alpha-q, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
7YFD
 
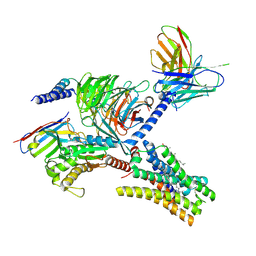 | | Cryo-EM structure of the imetit-bound histamine H4 receptor and Gq complex | | Descriptor: | 2-(1~{H}-imidazol-5-yl)ethyl carbamimidothioate, CHOLESTEROL, Engineered G-alpha-q, ... | | Authors: | Im, D, Iwata, S, Asada, H. | | Deposit date: | 2022-07-08 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the agonists binding and receptor selectivity of human histamine H 4 receptor.
Nat Commun, 14, 2023
|
|
1WWH
 
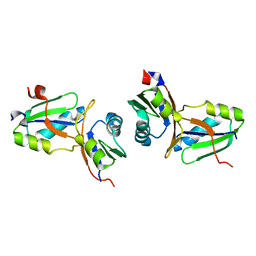 | | Crystal structure of the MPPN domain of mouse Nup35 | | Descriptor: | nucleoporin 35 | | Authors: | Handa, N, Murayama, K, Kukimoto, M, Hamana, H, Uchikubo, T, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-05 | | Release date: | 2005-07-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of mouse Nup35 reveals atypical RNP motifs and novel homodimerization of the RRM domain
J.Mol.Biol., 363, 2006
|
|
5XHF
 
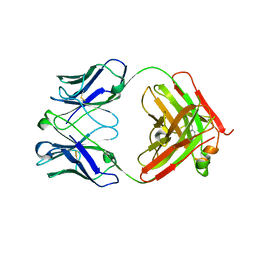 | | Crystal structure of Trastuzumab Fab fragment bearing p-azido-L-phenylalanine | | Descriptor: | polypeptide (H chain), polypeptide (L chain) | | Authors: | Kuratani, M, Yanagisawa, T, Sakamoto, K, Yokoyama, S. | | Deposit date: | 2017-04-20 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Extensive Survey of Antibody Invariant Positions for Efficient Chemical Conjugation Using Expanded Genetic Codes.
Bioconjug. Chem., 28, 2017
|
|
5XLI
 
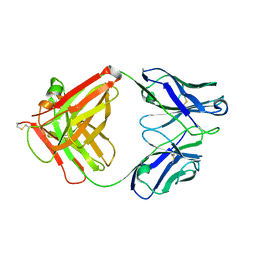 | | Structure of anti-Angiotensin II type2 receptor antibody (D5711-4A03) | | Descriptor: | FabH, FabL | | Authors: | Asada, H, Horita, S, Iwata, S, Hirata, K. | | Deposit date: | 2017-05-10 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Crystal structure of the human angiotensin II type 2 receptor bound to an angiotensin II analog.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3VJM
 
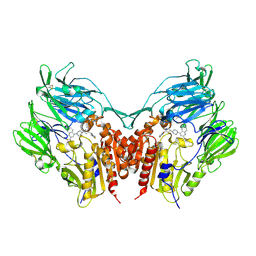 | | Crystal structure of human depiptidyl peptidase IV (DPP-4) in complex with a prolylthiazolidine inhibitor #1 | | Descriptor: | 1,3-thiazolidin-3-yl[(2S,4S)-4-{4-[2-(trifluoromethyl)quinolin-4-yl]piperazin-1-yl}pyrrolidin-2-yl]methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Akahoshi, F, Kishida, H, Miyaguchi, I, Yoshida, T, Ishii, S. | | Deposit date: | 2011-10-24 | | Release date: | 2012-08-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fused bicyclic heteroarylpiperazine-substituted l-prolylthiazolidines as highly potent DPP-4 inhibitors lacking the electrophilic nitrile group
Bioorg.Med.Chem., 20, 2012
|
|
1YCP
 
 | |
1OEL
 
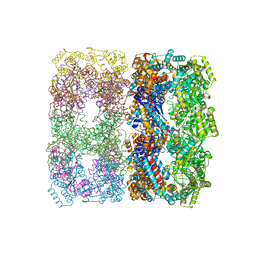 | |
5WT4
 
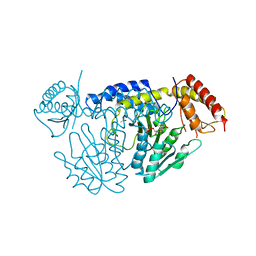 | | L-Cysteine-PLP intermediate of NifS from Helicobacter pylori | | Descriptor: | Cysteine desulfurase IscS, ISOPROPYL ALCOHOL, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-L-CYSTEINE | | Authors: | Fujishiro, T, Nakamura, R, Takahashi, T. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 2019
|
|
1KOE
 
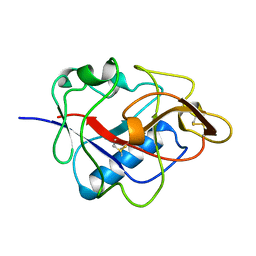 | | ENDOSTATIN | | Descriptor: | ENDOSTATIN | | Authors: | Hohenester, E, Sasaki, T, Olsen, B.R, Timpl, R. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-16 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the angiogenesis inhibitor endostatin at 1.5 A resolution.
EMBO J., 17, 1998
|
|
5YD8
 
 | |
1UL5
 
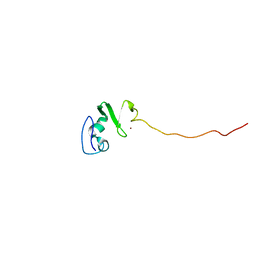 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 7 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 7 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
5YSD
 
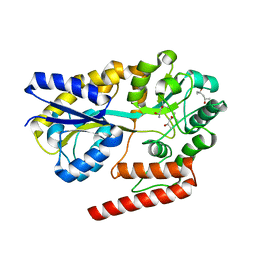 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotriose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSF
 
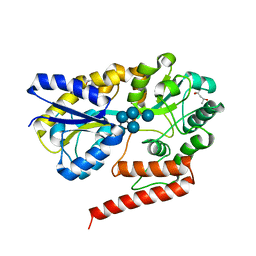 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophoropentaose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSE
 
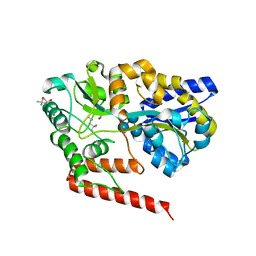 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
5YSB
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in ligand-free form | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lin1841 protein, ZINC ION | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-13 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
1UL4
 
 | | Solution structure of the DNA-binding domain of squamosa promoter binding protein-like 4 | | Descriptor: | ZINC ION, squamosa promoter binding protein-like 4 | | Authors: | Yamasaki, K, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-09 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors.
J.Mol.Biol., 337, 2004
|
|
