7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7ZFC
 
 | | SARS-CoV-2 Beta RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | Nanobody C1, Omi-18 heavy chain, Omi-18 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7AMT
 
 | | Structure of LuxR with DNA (activation) | | Descriptor: | DNA (5'-D(P*AP*TP*AP*AP*TP*GP*AP*CP*AP*TP*TP*AP*CP*TP*GP*TP*AP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*AP*GP*TP*AP*AP*TP*GP*TP*CP*AP*TP*TP*AP*T)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
7ZFB
 
 | | SARS-CoV-2 Omicron RBD in complex with nanobody C1, Omi-18 and Omi-31 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody C1, Omi-18 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF9
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with COVOX-150 Fab (P21) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-150 heavy chain, COVOX-150 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF7
 
 | | SARS-CoV-2 Omicron BA.2 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF5
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-12 and Beta-54 Fabs | | Descriptor: | Beta-54 heavy chain, Beta-54 light chain, Omi-12 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.32 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7ZF4
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-9 Fab and nanobody F2 | | Descriptor: | Nanobody F2, Omi-9 heavy chain, Omi-9 light chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
7Q1W
 
 | | Ruminococcus gnavus ATC29149 endo-beta-1,4-galactosidase (RgGH98) E411A in complex with blood group A (BgA II) tetrasaccharide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Owen, C.D, Wu, H, Crost, E.H, van Bakel, W, Gascuena, A.M, Latousakis, D, Hicks, T, Walpole, S, Urbanowicz, P.A, Ndeh, D, Monaco, S, Salom, L.S, Griffiths, R, Colvile, A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The human gut symbiont Ruminococcus gnavus shows specificity to blood group A antigen during mucin glycan foraging: Implication for niche colonisation in the gastrointestinal tract.
Plos Biol., 19, 2021
|
|
2C2W
 
 | |
7SGT
 
 | |
7ZXU
 
 | | SARS-CoV-2 Omicron BA.4/5 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 heavy chain, Beta-27 light chain, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-05-23 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum.
Cell, 185, 2022
|
|
7ZFA
 
 | | SARS-CoV-2 Omicron RBD in complex with Omi-6 and COVOX-150 Fabs | | Descriptor: | COVOX-150 heavy chain, COVOX-150 light chain, Omi-6 heavy chain, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (4.24 Å) | | Cite: | Potent cross-reactive antibodies following Omicron breakthrough in vaccinees.
Cell, 185, 2022
|
|
9CO2
 
 | |
1TPB
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
7AMZ
 
 | | Crystal structure of human Butyrylcholinesterase in complex with N-((2S,3R)-4-((2,2-dimethylpropyl)amino)-3-hydroxy-1-phenylbutan-2-yl)-2,2-diphenylacetamide | | Descriptor: | 2,2-dimethylpropyl-[(2~{R},3~{S})-3-(2,2-diphenylethanoylamino)-2-oxidanyl-4-phenyl-butyl]azanium, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Pasieka, A, Panek, D, Wieckowska, A. | | Deposit date: | 2020-10-10 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of multifunctional anti-Alzheimer's agents with a unique mechanism of action including inhibition of the enzyme butyrylcholinesterase and gamma-aminobutyric acid transporters.
Eur.J.Med.Chem., 218, 2021
|
|
1TPC
 
 | | OFFSET OF A CATALYTIC LESION BY A BOUND WATER SOLUBLE | | Descriptor: | PHOSPHOGLYCOLOHYDROXAMIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Zhang, Z, Sugio, S, Komives, E.A, Liu, K.D, Knowles, J.R, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-02-03 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for pseudoreversion of the E165D lesion by the secondary S96P mutation in triosephosphate isomerase depends on the positions of active site water molecules.
Biochemistry, 34, 1995
|
|
1TOX
 
 | | DIPHTHERIA TOXIN DIMER COMPLEXED WITH NAD | | Descriptor: | DIPHTHERIA TOXIN (DIMERIC), NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bell, C.E, Eisenberg, D. | | Deposit date: | 1995-10-06 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide.
Biochemistry, 35, 1996
|
|
1UHL
 
 | | Crystal structure of the LXRalfa-RXRbeta LBD heterodimer | | Descriptor: | (2E,4E)-11-METHOXY-3,7,11-TRIMETHYLDODECA-2,4-DIENOIC ACID, 10-mer peptide from Nuclear receptor coactivator 2, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, ... | | Authors: | Svensson, S, Ostberg, T, Jacobsson, M, Norstrom, C, Stefansson, K, Hallen, D, Johansson, I.C, Zachrisson, K, Ogg, D, Jendeberg, L. | | Deposit date: | 2003-07-03 | | Release date: | 2004-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the heterodimeric complex of LXRalpha and RXRbeta ligand-binding domains in a fully agonistic conformation
Embo J., 22, 2003
|
|
7AUX
 
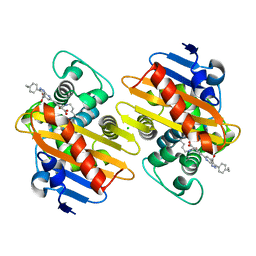 | | Crystal structure of OXA-48 beta-lactamase in the complex with the inhbitor ID2 | | Descriptor: | 6-(4-carboxyphenyl)-3-(4-ethylphenyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Beta-lactamase, CHLORIDE ION | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Garofalo, B, Ombrato, R. | | Deposit date: | 2020-11-03 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Novel Chemical Series of OXA-48 beta-Lactamase Inhibitors by High-Throughput Screening.
Pharmaceuticals, 14, 2021
|
|
2IQ7
 
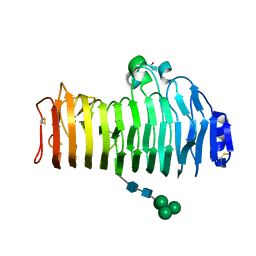 | | Crystal structure of the polygalacturonase from Colletotrichum lupini and its implications for the interaction with polygalacturonase-inhibiting proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bonivento, D, Federici, L, Matteo, A.D. | | Deposit date: | 2006-10-13 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the endopolygalacturonase from the phytopathogenic fungus Colletotrichum lupini and its interaction with polygalacturonase-inhibiting proteins
Proteins, 70, 2008
|
|
7AW5
 
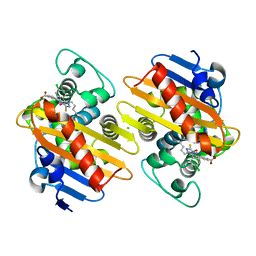 | | Crystal structure of OXA-48 beta-lactamase in the complex with the inhibitor ID3 | | Descriptor: | 4-[(~{E})-[3-(4-chlorophenyl)-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl]iminomethyl]benzoic acid, Beta-lactamase, CHLORIDE ION | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Garofalo, B, Ombrato, R. | | Deposit date: | 2020-11-06 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of Novel Chemical Series of OXA-48 beta-Lactamase Inhibitors by High-Throughput Screening.
Pharmaceuticals, 14, 2021
|
|
7QCX
 
 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, apo form | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
7QCY
 
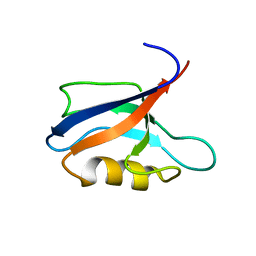 | | Two-state liquid NMR Structure of a PDZ2 Domain from hPTP1E, complexed with RA-GEF2 peptide | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 13 | | Authors: | Ashkinadze, D, Kadavath, H, Chi, C, Friedmann, M, Strotz, D, Kumari, P, Minges, M, Cadalbert, R, Koenigl, S, Guentert, P, Voegeli, B, Riek, R. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Nat Commun, 13, 2022
|
|
7AT5
 
 | | Structure of protein kinase ck2 catalytic subunit (csnk2a1 gene product) in complex with the bivalent inhibitor KN2 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,4-dichlorophenyl)ethanamine, Casein kinase II subunit alpha, ... | | Authors: | Lindenblatt, D, Applegate, V, Nickelsen, A, Klussmann, M, Neundorf, I, Goetz, C, Jose, J, Niefind, K. | | Deposit date: | 2020-10-29 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
J.Med.Chem., 65, 2022
|
|
