2JUV
 
 | | AbaA3-DKP-insulin | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Huang, K, Chan, S, Hua, Q, Chu, Y, Wang, R, Klaproth, B, Jia, W, Whittaker, J, De Meyts, P, Nakagawa, S.H, Steiner, D.F, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2007-09-05 | | Release date: | 2007-10-16 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | The A-chain of Insulin Contacts the Insert Domain of the Insulin Receptor: PHOTO-CROSS-LINKING AND MUTAGENESIS OF A DIABETES-RELATED CREVICE.
J.Biol.Chem., 282, 2007
|
|
2H95
 
 | |
2H7Z
 
 | | Crystal structure of irditoxin | | Descriptor: | Irditoxin subunit A, Irditoxin subunit B | | Authors: | Pawlak, J, Kini, R.M, Stura, E.A, Le Du, M.H. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-29 | | Last modified: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Irditoxin, a novel covalently linked heterodimeric three-finger toxin with high taxon-specific neurotoxicity.
Faseb J., 23, 2009
|
|
2J16
 
 | | Apo & Sulphate bound forms of SDP-1 | | Descriptor: | MAGNESIUM ION, SULFATE ION, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
2MD6
 
 | | NMR SOLUTION STRUCTURE OF ALPHA CONOTOXIN LO1A FROM Conus longurionis | | Descriptor: | ALPHA CONOTOXIN LO1A | | Authors: | Maiti, M, Lescrinier, E, Herdewijn, P, Lebbe, E.K.M, Peigneur, S, D'Souza, L, Tytgat, J. | | Deposit date: | 2013-09-01 | | Release date: | 2014-03-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure-Function Elucidation of a New alpha-Conotoxin, Lo1a, from Conus longurionis.
J.Biol.Chem., 289, 2014
|
|
4M51
 
 | | Crystal structure of amidohydrolase nis_0429 (ser145ala mutant) from nitratiruptor sp. sb155-2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Amidohydrolase family protein, BENZOIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Gobble, A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-07 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes.
Biochemistry, 52, 2013
|
|
2J17
 
 | | pTyr bound form of SDP-1 | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, TYROSINE-PROTEIN PHOSPHATASE YIL113W | | Authors: | Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2006-08-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Redox-mediated substrate recognition by Sdp1 defines a new group of tyrosine phosphatases.
Nature, 447, 2007
|
|
3V9E
 
 | | Structure of the L499M mutant of the laccase from B.aclada | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Osipov, E.M, Polyakov, K.M, Tikhonova, T.V, Dorovatovsky, P.V, Ludwig, R, Kittl, R, Shleev, S.V, Popov, V.O. | | Deposit date: | 2011-12-27 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Effect of the L499M mutation of the ascomycetous Botrytis aclada laccase on redox potential and catalytic properties.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q50
 
 | |
1FV3
 
 | | THE HC FRAGMENT OF TETANUS TOXIN COMPLEXED WITH AN ANALOGUE OF ITS GANGLIOSIDE RECEPTOR GT1B | | Descriptor: | ETHYL-TRIMETHYL-SILANE, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-beta-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Fotinou, C, Emsley, P, Black, I, Ando, H, Ishida, H, Kiso, M, Sinha, K.A, Fairweather, N.F, Isaacs, N.W. | | Deposit date: | 2000-09-18 | | Release date: | 2001-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of tetanus toxin Hc fragment complexed with a synthetic GT1b analogue suggests cross-linking between ganglioside receptors and the toxin.
J.Biol.Chem., 276, 2001
|
|
2I4W
 
 | | HIV-1 protease WT with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4U
 
 | | HIV-1 protease with TMC-126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
1OKK
 
 | |
4RCL
 
 | |
5YC9
 
 | |
3JXB
 
 | |
2OLA
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, cubic crystal form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Sauder, J.M, Ozyurt, S, Wasserman, S.R, Smith, D, Dickey, M, Maletic, M, Reyes, C, Gheyi, T, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OKT
 
 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, ligand-free form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Toro, R, Malashkevich, V, Sauder, J.M, Ozyurt, S, Smith, D, Dickey, M, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3KLN
 
 | | Vibrio cholerae VpsT | | Descriptor: | Transcriptional regulator, LuxR family | | Authors: | Krasteva, P.V, Navarro, V.A.S, Sondermann, H. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Vibrio cholerae VpsT Regulates Matrix Production and Motility by Directly Sensing Cyclic di-GMP.
Science, 327, 2010
|
|
3KLO
 
 | | Vibrio cholerae VpsT bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), D(-)-TARTARIC ACID, Transcriptional regulator VpsT | | Authors: | Krasteva, P.V, Navarro, V.A.S, Sondermann, H. | | Deposit date: | 2009-11-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Vibrio cholerae VpsT Regulates Matrix Production and Motility by Directly Sensing Cyclic di-GMP.
Science, 327, 2010
|
|
3JXC
 
 | |
3JXD
 
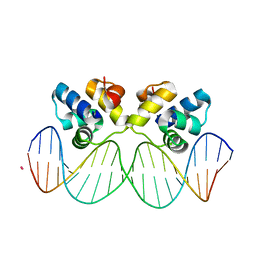 | |
2I4D
 
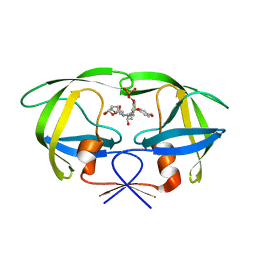 | | Crystal structure of WT HIV-1 protease with GS-8373 | | Descriptor: | ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONIC ACID, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-21 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4V
 
 | | HIV-1 protease I84V, L90M with TMC126 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]CARBAMATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
2I4X
 
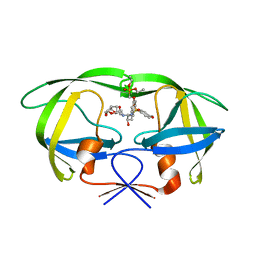 | | HIV-1 Protease I84V, L90M with GS-8374 | | Descriptor: | DIETHYL ({4-[(2S,3R)-2-({[(3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YLOXY]CARBONYL}AMINO)-3-HYDROXY-4-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}BUTYL]PHENOXY}METHYL)PHOSPHONATE, Protease | | Authors: | Hatada, M. | | Deposit date: | 2006-08-22 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Suppression of HIV-1 Protease Inhibitor Resistance by Phosphonate-mediated Solvent Anchoring.
J.Mol.Biol., 363, 2006
|
|
