2VYO
 
 | | Chitin deacetylase family member from Encephalitozoon cuniculi | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Urch, J.E, Hurtado-Guerrero, R, Texier, C, Van Aalten, D.M.F. | | Deposit date: | 2008-07-25 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of a Putative Polysaccharide Deacetylase of the Human Parasite Encephalitozoon Cuniculi.
Protein Sci., 18, 2009
|
|
1Q2W
 
 | | X-Ray Crystal Structure of the SARS Coronavirus Main Protease | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like protease | | Authors: | Bonanno, J.B, Fowler, R, Gupta, S, Hendle, J, Lorimer, D, Romero, R, Sauder, J.M, Wei, C.L, Liu, E.T, Burley, S.K, Harris, T. | | Deposit date: | 2003-07-26 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Company Says It Mapped Part of SARS Virus
New York Times, 30 July, 2003
|
|
2QR3
 
 | | Crystal structure of the N-terminal signal receiver domain of two-component system response regulator from Bacteroides fragilis | | Descriptor: | Two-component system response regulator | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Mendoza, M, Romero, R, Fong, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-27 | | Release date: | 2007-08-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the N-terminal signal receiver domain of two-component system response regulator from Bacteroides fragilis.
To be Published
|
|
2XUC
 
 | | Natural product-guided discovery of a fungal chitinase inhibitor | | Descriptor: | 1-methyl-3-(N-methylcarbamimidoyl)urea, CHITINASE, CHLORIDE ION, ... | | Authors: | Rush, C.L, Schuttelkopf, A.W, Hurtado-Guerrero, R, Blair, D.E, Ibrahim, A.F.M, Desvergnes, S, Eggleston, I.M, van Aalten, D.M.F. | | Deposit date: | 2010-10-18 | | Release date: | 2010-10-27 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Natural Product-Guided Discovery of a Fungal Chitinase Inhibitor.
Chem.Biol., 17, 2010
|
|
3DEC
 
 | | Crystal structure of a glycosyl hydrolases family 2 protein from Bacteroides thetaiotaomicron | | Descriptor: | Beta-galactosidase, POTASSIUM ION | | Authors: | Kumaran, D, Bonanno, J, Romero, R, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-09 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of a Glycosyl Hydrolases Family 2 protein from Bacteroides thetaiotaomicron.
To be Published
|
|
3DJC
 
 | | CRYSTAL STRUCTURE OF PANTOTHENATE KINASE FROM LEGIONELLA PNEUMOPHILA | | Descriptor: | GLYCEROL, Type III pantothenate kinase | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Dickey, M, Logan, C, Wasserman, S, Maletic, M, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-23 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantothenate Kinase from Legionella Pneumophila
To be Published
|
|
3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
3CS3
 
 | | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis | | Descriptor: | GLYCEROL, SULFATE ION, Sugar-binding transcriptional regulator, ... | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-08 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis.
To be Published
|
|
3CZ8
 
 | | Crystal structure of putative sporulation-specific glycosylase ydhD from Bacillus subtilis | | Descriptor: | GLYCEROL, Putative sporulation-specific glycosylase ydhD | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Chang, S, Maletic, M, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of putative glycosylase ydhD from Bacillus subtilis.
To be Published
|
|
3CTA
 
 | | Crystal structure of riboflavin kinase from Thermoplasma acidophilum | | Descriptor: | Riboflavin kinase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Mendoza, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of riboflavin kinase from Thermoplasma acidophilum.
To be Published
|
|
3CNB
 
 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
3CG0
 
 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
1T2X
 
 | | Glactose oxidase C383S mutant identified by directed evolution | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | Authors: | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
3FRN
 
 | | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima MSB8 | | Descriptor: | Flagellar protein FlgA, GLYCEROL | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Hu, S, Bain, K, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CRYSTAL STRUCTURE OF flagellar protein FlgA FROM Thermotoga maritima
To be Published
|
|
3DLS
 
 | | Crystal structure of human PAS kinase bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PAS domain-containing serine/threonine-protein kinase | | Authors: | Antonysamy, S, Bonanno, J.B, Romero, R, Russell, M, Iizuka, M, Gheyi, T, Wasserman, S.R, Rutter, J, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-29 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural bases of PAS domain-regulated kinase (PASK) activation in the absence of activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
1TEY
 
 | | NMR structure of human histone chaperone, ASF1A | | Descriptor: | ASF1 anti-silencing function 1 homolog A | | Authors: | Mousson, F, Lautrette, A, Thuret, J.Y, Agez, M, Amigues, B, Courbeyrette, R, Neumann, J.M, Guerois, R, Mann, C, Ochsenbein, F. | | Deposit date: | 2004-05-26 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of Asf1 with histone H3 and its functional implications.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1L7B
 
 | | Solution NMR Structure of BRCT Domain of T. Thermophilus: Northeast Structural Genomics Consortium Target WR64TT | | Descriptor: | DNA LIGASE | | Authors: | Sahota, G, Dixon, B.L, Huang, Y.P, Aramini, J, Monleon, D, Bhattacharya, D, Swapna, G.V.T, Yin, C, Xiao, R, Anderson, S, Tejero, R, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-14 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Brct Domain from Thermus Thermophilus DNA Ligase
To be Published
|
|
1P45
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B.N, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
1NS1
 
 | |
2YGV
 
 | | Conserved N-terminal domain of the yeast Histone Chaperone Asf1 in complex with the C-terminal fragment of Rad53 | | Descriptor: | GLYCEROL, HISTONE CHAPERONE ASF1, SERINE/THREONINE-PROTEIN KINASE RAD53, ... | | Authors: | Jiao, Y, Seeger, K, Murciano, B, Ledu, M.H, Charbonnier, J.B, Legrand, P, Lautrette, A, Gaubert, A, Mousson, F, Guerois, R, Mann, C, Ochsenbein, F. | | Deposit date: | 2011-04-21 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Surprising Complexity of the Asf1 Histone Chaperone-Rad53 Kinase Interaction
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2XTB
 
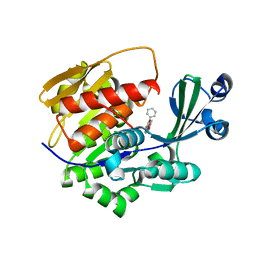 | | Crystal Structure of Trypanosoma brucei rhodesiense Adenosine Kinase Complexed with Activator | | Descriptor: | 4-[5-(4-PHENOXYPHENYL)-1H-PYRAZOL-3-YL]MORPHOLINE, ADENOSINE KINASE | | Authors: | Kuettel, S, Greenwald, J, Kostrewa, D, Ahmed, S, Scapozza, L, Perozzo, R. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of T. B. Rhodesiense Adenosine Kinase Complexed with Inhibitor and Activator: Implications for Catalysis and Hyperactivation.
Plos Negl Trop Dis, 5, 2011
|
|
1Z6B
 
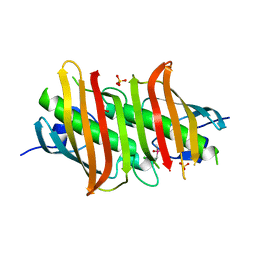 | | Crystal structure of Plasmodium falciparum FabZ at 2.1 A | | Descriptor: | CACODYLATE ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kostrewa, D, Winkler, F.K, Folkers, G, Scapozza, L, Perozzo, R. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum
PROTEIN SCI., 14, 2005
|
|
1P44
 
 | | Targeting tuberculosis and malaria through inhibition of enoyl reductase: compound activity and structural data | | Descriptor: | 5-{[4-(9H-FLUOREN-9-YL)PIPERAZIN-1-YL]CARBONYL}-1H-INDOLE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kuo, M.R, Morbidoni, H.R, Alland, D, Sneddon, S.F, Gourlie, B.B, Staveski, M.M, Leonard, M, Gregory, J.S, Janjigian, A.D, Yee, C, Musser, J.M, Kreiswirth, B, Iwamoto, H, Perozzo, R, Jacobs Jr, W.R, Sacchettini, J.C, Fidock, D.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-04-21 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting tuberculosis and malaria through inhibition of Enoyl reductase: compound activity and structural data.
J.Biol.Chem., 278, 2003
|
|
3BHW
 
 | | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum | | Descriptor: | Uncharacterized protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an uncharacterized protein from Magnetospirillum magneticum.
To be Published
|
|
3BGH
 
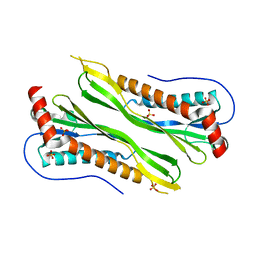 | | Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori | | Descriptor: | Putative neuraminyllactose-binding hemagglutinin homolog, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, J, Bain, K.T, McKenzie, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-26 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of putative neuraminyllactose-binding hemagglutinin homolog from Helicobacter pylori.
To be Published
|
|
