7K86
 
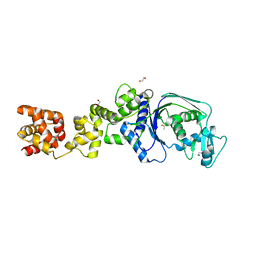 | |
7N4D
 
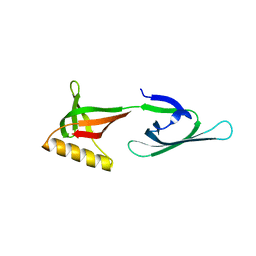 | |
7L9R
 
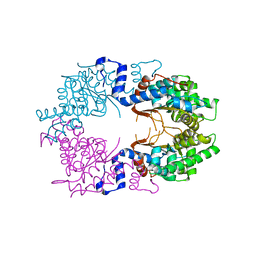 | |
1TFG
 
 | |
2SOD
 
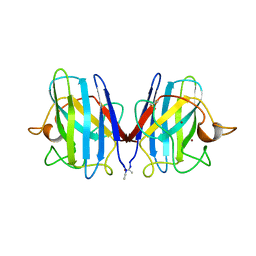 | | DETERMINATION AND ANALYSIS OF THE 2 ANGSTROM STRUCTURE OF COPPER, ZINC SUPEROXIDE DISMUTASE | | Descriptor: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Tainer, J.A, Getzoff, E.D, Richardson, J.S, Richardson, D.C. | | Deposit date: | 1980-03-25 | | Release date: | 1980-05-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase.
J.Mol.Biol., 160, 1982
|
|
1PII
 
 | |
4JE1
 
 | |
4HWG
 
 | |
1AGQ
 
 | | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR FROM RAT | | Descriptor: | GLIAL CELL-DERIVED NEUROTROPHIC FACTOR | | Authors: | Eigenbrot, C, Gerber, N. | | Deposit date: | 1997-03-25 | | Release date: | 1997-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of glial cell-derived neurotrophic factor at 1.9 A resolution and implications for receptor binding.
Nat.Struct.Biol., 4, 1997
|
|
1BHL
 
 | | CACODYLATED CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-10 | | Release date: | 1998-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1BI4
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-06-22 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
1C29
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-23 | | Release date: | 2000-01-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1BL3
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
5ER1
 
 | |
1DMA
 
 | | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH NICOTINAMIDE AND AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, EXOTOXIN A, NICOTINAMIDE | | Authors: | Li, M, Dyda, F, Benhar, I, Pastan, I, Davies, D. | | Deposit date: | 1995-04-28 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of Pseudomonas aeruginosa exotoxin domain III with nicotinamide and AMP: conformational differences with the intact exotoxin.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
7MFC
 
 | |
6MBQ
 
 | | Crystal structure of Mg-free wild-type KRAS (2-166) bound to GMPPNP in the state 1 conformation | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Dharmaiah, S, Davies, D.R, Abendroth, J, Gillette, W.G, Stephen, A.G, Simanshu, D.K. | | Deposit date: | 2018-08-30 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of N-terminally processed KRAS provide insight into the role of N-acetylation.
Sci Rep, 9, 2019
|
|
1DKK
 
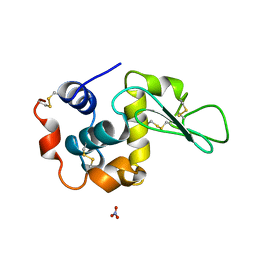 | | BOBWHITE QUAIL LYSOZYME WITH NITRATE | | Descriptor: | LYSOZYME, NITRATE ION | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
1DKJ
 
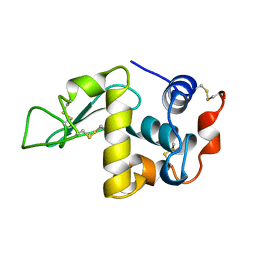 | | BOBWHITE QUAIL LYSOZYME | | Descriptor: | LYSOZYME | | Authors: | Jeffrey, P.D, Sheriff, S. | | Deposit date: | 1996-01-10 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures of bobwhite quail lysozyme uncomplexed and complexed with the HyHEL-5 Fab fragment.
Proteins, 26, 1996
|
|
1XC4
 
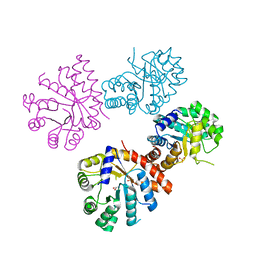 | |
4APE
 
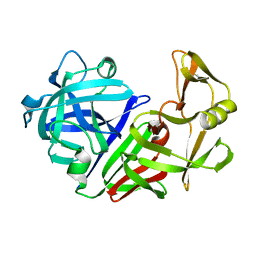 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | ENDOTHIAPEPSIN | | Authors: | Pearl, L.H, Sewell, B.T, Jenkins, J.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1986-06-09 | | Release date: | 1986-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
2ITG
 
 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE: ORDERED ACTIVE SITE IN THE F185H CONSTRUCT | | Descriptor: | HUMAN IMMUNODEFICIENCY VIRUS-1 INTEGRASE | | Authors: | Bujacz, G, Alexandratos, J, Wlodawer, A, Zhou-Liu, Q, Clement-Mella, C. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant.
FEBS Lett., 398, 1996
|
|
1CX9
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-AMINOPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-AMINOPHENYLTHIO)-BUTYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-08-29 | | Release date: | 1999-12-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
1ER8
 
 | | THE ACTIVE SITE OF ASPARTIC PROTEINASES | | Descriptor: | Endothiapepsin, H-77 | | Authors: | Hemmings, A.M, Veerapandian, B, Szelke, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1989-10-16 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Active Site of Aspartic Proteinases
FEBS Lett., 174, 1984
|
|
