6SPO
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SPQ
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase variant VI298 complexed with methionine | | Descriptor: | CITRIC ACID, GLYCEROL, METHIONINE, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6SW9
 
 | | IC2A model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
7ZKI
 
 | | Cryo-EM structure of aIF1A:aIF5B:Met-tRNAiMet complex from a Pyrococcus abyssi 30S initiation complex | | Descriptor: | MAGNESIUM ION, METHIONINE, Met-tRNAiMet, ... | | Authors: | Coureux, P.D, Bourgeois, G, Mechulam, Y, Schmitt, E, Kazan, R. | | Deposit date: | 2022-04-13 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
6SPN
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
6L1D
 
 | |
6L1M
 
 | |
3V11
 
 | |
1BOE
 
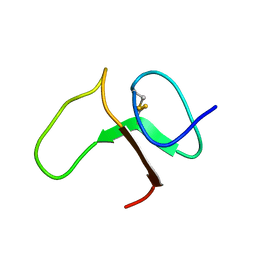 | | STRUCTURE OF THE IGF BINDING DOMAIN OF THE INSULIN-LIKE GROWTH FACTOR-BINDING PROTEIN-5 (IGFBP-5): IMPLICATIONS FOR IGF AND IGF-I RECEPTOR INTERACTIONS | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR-BINDING PROTEIN-5 (IGFBP-5)) | | Authors: | Kalus, W, Zweckstetter, M, Renner, C, Sanchez, Y, Georgescu, J, Grol, M, Demuth, D, Schumacherdony, C, Lang, K, Holak, T.H. | | Deposit date: | 1998-07-30 | | Release date: | 1998-12-16 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure of the IGF-binding domain of the insulin-like growth factor-binding protein-5 (IGFBP-5): implications for IGF and IGF-I receptor interactions.
EMBO J., 17, 1998
|
|
3NCU
 
 | |
8Q95
 
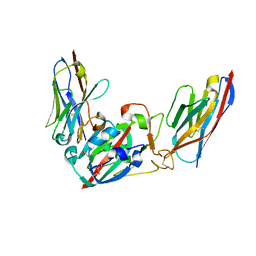 | | Crystal structure of the SARS-CoV-2 BA.1 RBD with neutralizing-VHHs Ma16B06 and Ma3F05 | | Descriptor: | Nanobody Ma16B06, Nanobody Ma3F05, Spike protein S1 | | Authors: | Aksu, M, Rymarenko, O, Guttler, T, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q94
 
 | |
8Q7S
 
 | | Crystal structure of the SARS-CoV-2 RBD (Wuhan) with neutralizing VHHs Ma6F06 and Re21H01 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, GLYCEROL, ... | | Authors: | Guttler, T, Aksu, M, Gorlich, D. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q93
 
 | | Crystal structure of the SARS-COV-2 RBD with neutralizing-VHHs Re30H02 and Re21D01 | | Descriptor: | Nanobody Re21D01, Nanobody Re30H02, Spike protein S1 | | Authors: | Aksu, M, Guttler, T, Rymarenko, O, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
6P6G
 
 | | Co-crystal Structure of human SMYD3 with Isoxazole Amides Inhibitors | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6P6K
 
 | |
6P7Z
 
 | |
6PAF
 
 | |
6QMO
 
 | |
6QN4
 
 | |
5U3I
 
 | | CRYSTAL STRUCTURE OF CARBONMONOXY HEMOGLOBIN S (LIGANDED SICKLE CELL HEMOGLOBIN) COMPLEXED WITH GBT compound 31 | | Descriptor: | 2-methoxy-5-({2-[1-(propan-2-yl)-1H-pyrazol-5-yl]pyridin-3-yl}methoxy)pyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2016-12-02 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
5UFJ
 
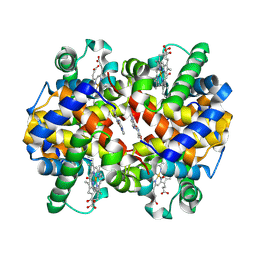 | | Crystal Structure of Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Complexed with GBT Compound 6 | | Descriptor: | 5-[(imidazo[1,2-a]pyridin-8-yl)methoxy]-2-methoxypyridine-4-carbaldehyde, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Partridge, J.R, Choy, R.M, Li, Z, Metcalf, B. | | Deposit date: | 2017-01-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of GBT440, an Orally Bioavailable R-State Stabilizer of Sickle Cell Hemoglobin.
ACS Med Chem Lett, 8, 2017
|
|
6ZH0
 
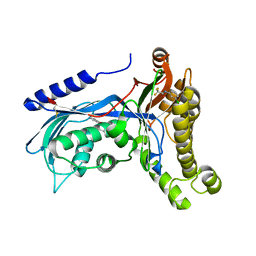 | | Structure of human galactokinase 1 bound with 2-(4-chlorophenyl)-N-(pyrimidin-2-yl)acetamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(3-chlorophenyl)-2,2,2-trifluoroacetamide, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2020-06-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment Screening Reveals Starting Points for Rational Design of Galactokinase 1 Inhibitors to Treat Classic Galactosemia.
Acs Chem.Biol., 16, 2021
|
|
4NYF
 
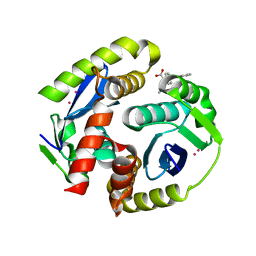 | | HIV integrase in complex with inhibitor | | Descriptor: | (2S)-tert-butoxy[4-(4-chlorophenyl)-2-methylquinolin-3-yl]ethanoic acid, CADMIUM ION, Integrase | | Authors: | Coulombe, R, Fader, L. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of BI 224436, a Noncatalytic Site Integrase Inhibitor (NCINI) of HIV-1.
ACS Med Chem Lett, 5, 2014
|
|
8PX3
 
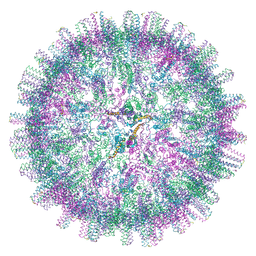 | |
