4KMY
 
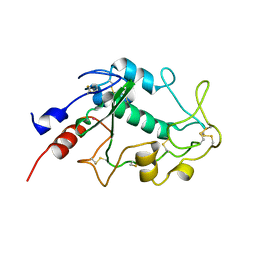 | | Human folate receptor beta (FOLR2) at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Folate receptor beta, POTASSIUM ION | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KM7
 
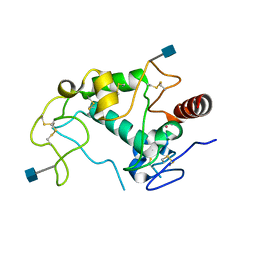 | | Human folate receptor alpha (FOLR1) at acidic pH, triclinic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Folate receptor alpha, POTASSIUM ION | | Authors: | Kovach, A.R, Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KM6
 
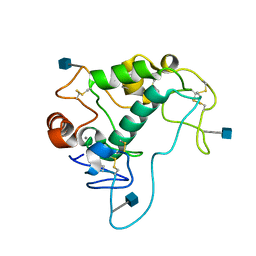 | | Human folate receptor alpha (FOLR1) at acidic pH, orthorhombic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Folate receptor alpha | | Authors: | Singh, M, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KN1
 
 | |
4KMZ
 
 | | Human folate receptor beta (FOLR2) in complex with the folate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, FOLIC ACID, ... | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KN2
 
 | | Human folate receptor beta (FOLR2) in complex with antifolate pemetrexed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, CHLORIDE ION, ... | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8J5D
 
 | | Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH | | Descriptor: | Beta-amylase 1, chloroplastic, Malate dehydrogenase, ... | | Authors: | Guan, Z.Y, Liu, J, Yan, J.J. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Plant Cell, 36, 2023
|
|
8JGR
 
 | |
8JGT
 
 | |
8JGX
 
 | |
8JGW
 
 | |
8JGU
 
 | |
8JGQ
 
 | |
8JGP
 
 | |
8JGO
 
 | |
7XT6
 
 | | Structure of a membrane protein M3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell antigen receptor complex-associated protein alpha chain, ... | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-05-16 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
5YS3
 
 | |
5YS8
 
 | |
6IUF
 
 | | Crystal structure of Anti-CRISPR protein AcrVA5 | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, protein-a | | Authors: | Dong, L, Guan, X, Zhu, Y, Huang, Z. | | Deposit date: | 2018-11-28 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | An anti-CRISPR protein disables type V Cas12a by acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6IBI
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
6IV6
 
 | | Cryo-EM structure of AcrVA5-acetylated MbCas12a in complex with crRNA | | Descriptor: | RNA (59-MER), nuclease | | Authors: | Dong, L, Li, N, Guan, X, Zhu, Y, Gao, N, Huang, Z. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An anti-CRISPR protein disables type V Cas12a by acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
6IBJ
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
6IBH
 
 | | Copper binding protein from Laetisaria arvalis (LaX325) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity CAZyme, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Tandrup, T, Labourel, A, Haon, M, Berrin, J.-G, Lo Leggio, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-11-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fungal family of lytic polysaccharide monooxygenase-like copper proteins.
Nat.Chem.Biol., 16, 2020
|
|
6KRG
 
 | | Crystal structure of sfGFP Y182TMSiPhe | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
4OO8
 
 | | Crystal structure of Streptococcus pyogenes Cas9 in complex with guide RNA and target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*CP*AP*GP*CP*CP*AP*AP*GP*CP*GP*CP*AP*CP*CP*TP*AP*AP*TP*TP*TP*CP*C)-3'), RNA (97-MER) | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2014-01-31 | | Release date: | 2014-02-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Cas9 in complex with guide RNA and target DNA
Cell(Cambridge,Mass.), 156, 2014
|
|
