3ZG5
 
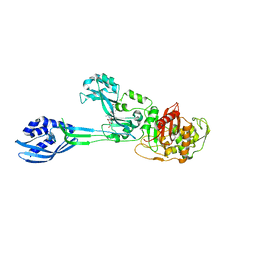 | | Crystal structure of PBP2a from MRSA in complex with peptidoglycan analogue at allosteric | | Descriptor: | CADMIUM ION, CHLORIDE ION, PEPTIDOGLYCAN ANALOGUE, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2012-12-14 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How Allosteric Control of Staphylococcus Aureus Penicillin Binding Protein 2A Enables Methicillin Resistance and Physiological Function
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7LQV
 
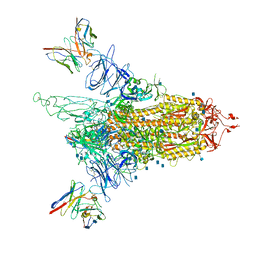 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
7LQW
 
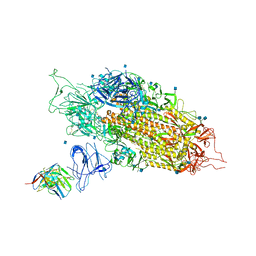 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | Descriptor: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
5OAU
 
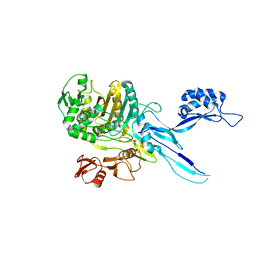 | |
6JN8
 
 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
5OJ1
 
 | | Penicillin Binding Protein 2x (PBP2x) from S.pneumoniae in complex with Oxacillin and a tetrasaccharide | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X, SODIUM ION | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
6JMZ
 
 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
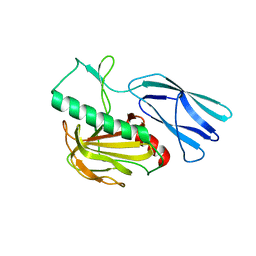 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
5OIZ
 
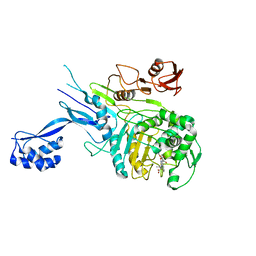 | | Penicillin-Binding Protein 2X (PBP2X) from Streptococcus pneumoniae in complex with oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 2X | | Authors: | Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allostery, Recognition of Nascent Peptidoglycan, and Cross-linking of the Cell Wall by the Essential Penicillin-Binding Protein 2x of Streptococcus pneumoniae.
ACS Chem. Biol., 13, 2018
|
|
2JTN
 
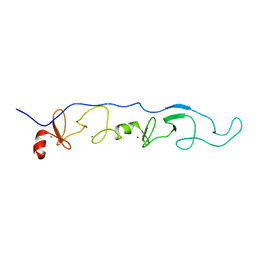 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | Descriptor: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | Authors: | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2007-08-03 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
6JN0
 
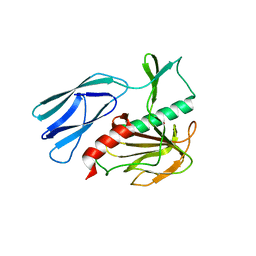 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
 | | Structure of wild type closed form of peptidoglycan peptidase | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
5OJ0
 
 | |
6KBM
 
 | | Crystal structure of Vac8 bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6E2M
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-methyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2N
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl][3,4'-bipyridine]-2'-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2O
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | 4-(4-cyclopropyl-1H-imidazol-1-yl)-N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
4K91
 
 | | Crystal structure of Penicillin-Binding Protein 5 (PBP5) from Pseudomonas aeruginosa in apo state | | Descriptor: | D-ala-D-ala-carboxypeptidase, SUCCINIC ACID | | Authors: | Smith, J, Toth, M, Vakulenko, S, Mobashery, S, Chen, Y. | | Deposit date: | 2013-04-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in beta-lactam resistance.
Antimicrob.Agents Chemother., 57, 2013
|
|
2J8G
 
 | |
2J8F
 
 | |
2IXU
 
 | |
6I0N
 
 | |
6I05
 
 | |
