5ZRD
 
 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at low pH condition | | Descriptor: | CITRIC ACID, COPPER (II) ION, GLYCEROL, ... | | Authors: | Lee, S, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
5ZRE
 
 | | Tyrosinase from Burkholderia thailandensis (BtTYR) at high pH condition | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN ATOM, ... | | Authors: | Lee, S, Son, H.-F, Kim, K.-J. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Highly Efficient Production of Catechol Derivatives at Acidic pH by Tyrosinase from Burkholderia thailandensis
Acs Catalysis, 8, 2018
|
|
5ZHF
 
 | | Structure of VanYB unbound | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-03-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
6A6A
 
 | | VanYB in complex with D-Alanine | | Descriptor: | ACETATE ION, D-ALANINE, D-alanyl-D-alanine carboxypeptidase, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
5ZHW
 
 | | VanYB in complex with D-Alanine-D-Alanine | | Descriptor: | COPPER (II) ION, D-ALANINE, D-alanyl-D-alanine carboxypeptidase, ... | | Authors: | Kim, H.S, Hahn, H. | | Deposit date: | 2018-03-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for the substrate recognition of peptidoglycan pentapeptides by Enterococcus faecalis VanYB.
Int. J. Biol. Macromol., 119, 2018
|
|
1Y4U
 
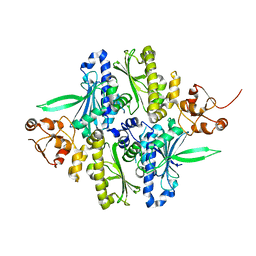 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
1Y4S
 
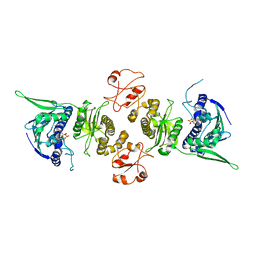 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
7LFO
 
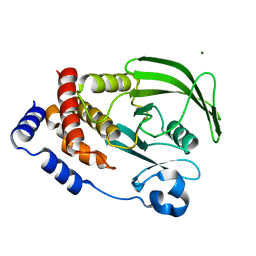 | | Protein Tyrosine Phosphatase 1B | | Descriptor: | MAGNESIUM ION, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Sarkar, A, Kim, E.Y, Hongdusit, A, Sankaran, B, Fox, J.M. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Microbially Guided Discovery and Biosynthesis of Biologically Active Natural Products.
Acs Synth Biol, 10, 2021
|
|
7EC1
 
 | | Crystal structure of SdgB (ligand-free form) | | Descriptor: | GLYCEROL, Glycosyl transferase, group 1 family protein, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC7
 
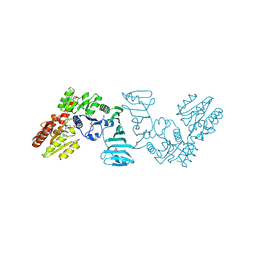 | | Crystal structure of SdgB (complexed with phosphate ions) | | Descriptor: | Glycosyl transferase, group 1 family protein, PHOSPHATE ION | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC6
 
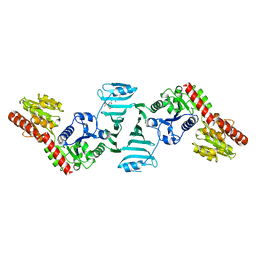 | | Crystal structure of SdgB (complexed with peptides) | | Descriptor: | ASP-SER-ASP, Glycosyl transferase, group 1 family protein | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EC3
 
 | | Crystal structure of SdgB (complexed with UDP, GlcNAc, and Glycosylated peptide) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-35)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-65)]5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyl transferase, ... | | Authors: | Kim, D.-G, Baek, I, Lee, Y, Kim, H.S. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SdgB- and SdgA-mediated glycosylation of staphylococcal adhesive proteins.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
3WUT
 
 | | Structure basis of inactivating cell abscission | | Descriptor: | Centrosomal protein of 55 kDa, GLYCEROL, Inactive serine/threonine-protein kinase TEX14 | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUV
 
 | | Structure basis of inactivating cell abscission with chimera peptide 2 | | Descriptor: | Centrosomal protein of 55 kDa, peptide from Programmed cell death 6-interacting protein | | Authors: | Kim, H.J, Matsuura, A, Lee, H.H. | | Deposit date: | 2014-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3WUU
 
 | |
5YSS
 
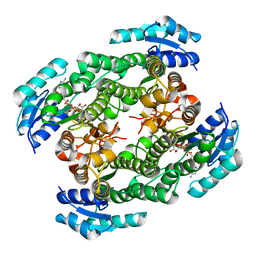 | |
5Y86
 
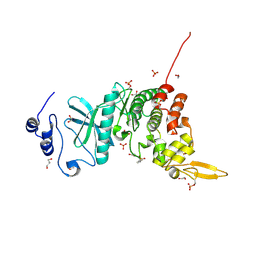 | | Crystal structure of kinase | | Descriptor: | 1,2-ETHANEDIOL, 7-METHOXY-1-METHYL-9H-BETA-CARBOLINE, Dual specificity tyrosine-phosphorylation-regulated kinase 3, ... | | Authors: | Kim, K.L, Cha, J.S, Cho, Y.S, Kim, H.Y, Chang, N.P, Cho, H.S. | | Deposit date: | 2017-08-18 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Dual-Specificity Tyrosine-Regulated Kinase 3 Reveals New Structural Features and Insights into its Auto-phosphorylation
J. Mol. Biol., 430, 2018
|
|
3P2F
 
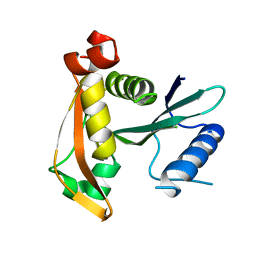 | | Crystal structure of TofI in an apo form | | Descriptor: | AHL synthase | | Authors: | Yu, S, Rhee, S. | | Deposit date: | 2010-10-02 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5ZXB
 
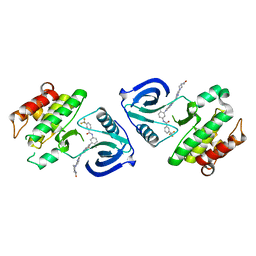 | | Crystal structure of ACK1 with compound 10d | | Descriptor: | Activated CDC42 kinase 1, N-{3-[7-{[6-(4-acetylpiperazin-1-yl)pyridin-3-yl]amino}-1-methyl-2-oxo-1,4-dihydropyrimido[4,5-d]pyrimidin-3(2H)-yl]-4-methylphenyl}-3-(trifluoromethyl)benzamide | | Authors: | Hong, E.M, Kim, H.L, Sim, T.B. | | Deposit date: | 2018-05-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | First SAR Study for Overriding NRAS Mutant Driven Acute Myeloid Leukemia.
J. Med. Chem., 61, 2018
|
|
3P2H
 
 | |
4O6G
 
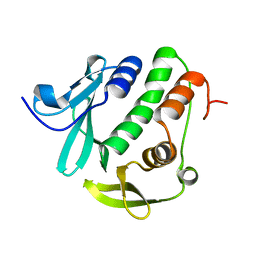 | | Rv3902c from M. tuberculosis | | Descriptor: | Uncharacterized protein | | Authors: | Reddy, B.G, Moates, D.B, Kim, H, Green, T.J, Kim, C, Terwilliger, T.J, Delucas, L.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 angstrom resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
6L3A
 
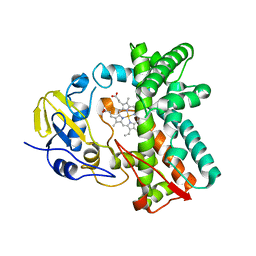 | | Cytochrome P450 107G1 (RapN) with everolimus | | Descriptor: | Cytochrome P450, Everolimus, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Km, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
7DIB
 
 | |
5Z59
 
 | | Crystal structure of Tk-PTP in the inactive form | | Descriptor: | Protein-tyrosine phosphatase | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
6YEJ
 
 | | Cryo-EM structure of the Full-length disease type human Huntingtin | | Descriptor: | Huntingtin | | Authors: | Tame, G, Jung, T, Dal Perraro, M, Hebert, H, Song, J. | | Deposit date: | 2020-03-24 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (18.200001 Å) | | Cite: | The Polyglutamine Expansion at the N-Terminal of Huntingtin Protein Modulates the Dynamic Configuration and Phosphorylation of the C-Terminal HEAT Domain.
Structure, 28, 2020
|
|
