5C3T
 
 | |
6YI4
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | 著者 | Zak, K.M, Zhou, R.X, Softley, C.A, Bostock, M.J, Sattler, M, Popowicz, G.M. | | 登録日 | 2020-03-31 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of IMP-13 metallo-beta-lactamase complexed with citrate anion
Not published
|
|
7OD0
 
 | | Mirolysin in complex with compound 9 | | 分子名称: | 1,2-ETHANEDIOL, 2,1,3-benzothiadiazol-4-ylmethanamine, ACETATE ION, ... | | 著者 | Zak, K.M, Bostock, M.J, Ksiazek, M. | | 登録日 | 2021-04-28 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Latency, thermal stability, and identification of an inhibitory compound of mirolysin, a secretory protease of the human periodontopathogen Tannerella forsythia .
J Enzyme Inhib Med Chem, 36, 2021
|
|
5J89
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | 分子名称: | 1,2-ETHANEDIOL, N-{2-[({2-methoxy-6-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]pyridin-3-yl}methyl)amino]ethyl}acetamide, Programmed cell death 1 ligand 1 | | 著者 | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | 登録日 | 2016-04-07 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
5J8O
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | 分子名称: | (2R)-1-({3-bromo-4-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]phenyl}methyl)piperidine-2-carboxylic acid, Programmed cell death 1 ligand 1 | | 著者 | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | 登録日 | 2016-04-08 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
4ZQK
 
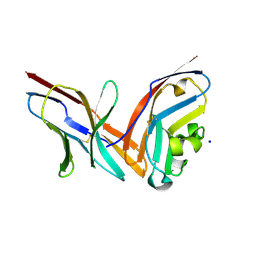 | | Structure of the complex of human programmed death-1 (PD-1) and its ligand PD-L1. | | 分子名称: | Programmed cell death 1 ligand 1, Programmed cell death protein 1, SODIUM ION | | 著者 | Zak, K.M, Dubin, G, Holak, T.A. | | 登録日 | 2015-05-10 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
8R3F
 
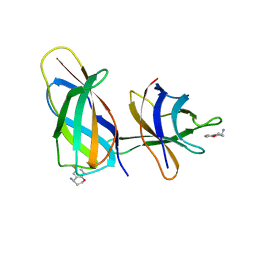 | | C-terminal Rel-homology Domain of NFAT1 | | 分子名称: | (4~{S})-6-fluoranyl-3,4-dihydro-2~{H}-chromen-4-amine, Nuclear factor of activated T-cells, cytoplasmic 2 | | 著者 | Zak, K.M, Boettcher, J. | | 登録日 | 2023-11-08 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Ligandability assessment of the C-terminal Rel-homology domain of NFAT1.
Arch Pharm, 357, 2024
|
|
8R07
 
 | |
5NIU
 
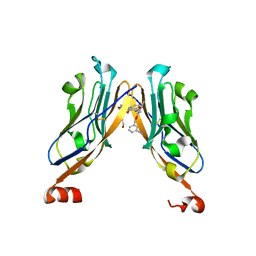 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | 分子名称: | (2~{R})-2-[[2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-5-methyl-phenyl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | 著者 | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | 登録日 | 2017-03-27 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells.
Oncotarget, 8, 2017
|
|
6RZR
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed imipenem | | 分子名称: | (2R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-(2-methanimidamidoethylsulfanyl)-2,3-dihydro-1H-pyrrole -5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | 登録日 | 2019-06-13 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6RZS
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed ertapenem | | 分子名称: | Beta-lactamase, ZINC ION, hydrolysed ertapenem | | 著者 | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | 登録日 | 2019-06-13 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6S0H
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed doripenem | | 分子名称: | (2~{R},3~{R})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-3-methyl-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-2,3-dihydro-1~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | 登録日 | 2019-06-14 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6R78
 
 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop closed) | | 分子名称: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Beta-lactamase, ... | | 著者 | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | 登録日 | 2019-03-28 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6R79
 
 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop open) | | 分子名称: | BETA-MERCAPTOETHANOL, Beta-lactamase, GLYCEROL, ... | | 著者 | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | 登録日 | 2019-03-28 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
4ZFI
 
 | | Structure of Mdm2 with low molecular weight inhibitor | | 分子名称: | (5S)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxy-1-methyl-1,5-dihydro-2H-pyrrol-2-one, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Zak, K.M, Twarda-Clapa, A, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2015-04-21 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
7NDX
 
 | | Crystal structure of the human HSP40 DNAJB1-CTDs in complex with a peptide of NudC | | 分子名称: | 1,2-ETHANEDIOL, DnaJ homolog subfamily B member 1, Nuclear migration protein nudC | | 著者 | Delhommel, F, Zak, K.M, Popowicz, G.M, Sattler, M. | | 登録日 | 2021-02-02 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.541 Å) | | 主引用文献 | NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems.
Mol.Cell, 82, 2022
|
|
5N2F
 
 | | Structure of PD-L1/small-molecule inhibitor complex | | 分子名称: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | 著者 | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2017-02-07 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
5N2D
 
 | | Structure of PD-L1/small-molecule inhibitor complex | | 分子名称: | Programmed cell death 1 ligand 1, ~{N}-[2-[[2,6-dimethoxy-4-[(2-methyl-3-phenyl-phenyl)methoxy]phenyl]methylamino]ethyl]ethanamide | | 著者 | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2017-02-07 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
4ZGK
 
 | | Structure of Mdm2 with low molecular weight inhibitor. | | 分子名称: | (5R)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxyfuran-2(5H)-one, E3 ubiquitin-protein ligase Mdm2 | | 著者 | Twarda-Clapa, A, Zak, K.M, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | 登録日 | 2015-04-23 | | 公開日 | 2016-10-19 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
6R2N
 
 | | Crystal structure of KlGlk1 glucokinase from Kluyveromyces lactis | | 分子名称: | 1,2-ETHANEDIOL, BROMIDE ION, Glucokinase-1 | | 著者 | Zak, K, Wator, E, Grudnik, P. | | 登録日 | 2019-03-18 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.596 Å) | | 主引用文献 | Crystal Structure of Kluyveromyces lactis Glucokinase ( Kl Glk1).
Int J Mol Sci, 20, 2019
|
|
6R3K
 
 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | 分子名称: | (2~{S},4~{R})-1-[[5-chloranyl-2-[(3-cyanophenyl)methoxy]-4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, Programmed cell death 1 ligand 1 | | 著者 | Zak, K.M, Grudnik, P, Skalniak, L, Dubin, G, Holak, T.A. | | 登録日 | 2019-03-20 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Terphenyl-Based Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7Z0J
 
 | | human PEX13 SH3 domain in complex with internal FxxxF motif | | 分子名称: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13 | | 著者 | Gaussmann, S, Zak, K, Kreisz, N, Sattler, M. | | 登録日 | 2022-02-23 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Intramolecular autoinhibition of human PEX13 modulates peroxisomal import
Biorxiv, 2022
|
|
7Z0I
 
 | | human PEX13 SH3 domain | | 分子名称: | 1,2-ETHANEDIOL, Peroxisomal membrane protein PEX13, ZINC ION | | 著者 | Gaussmann, S, Zak, K, Sattler, M. | | 登録日 | 2022-02-23 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Intramolecular autoinhibition of human PEX13 modulates peroxisomal import
Biorxiv, 2022
|
|
6R73
 
 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed meropenem | | 分子名称: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, ZINC ION | | 著者 | Softley, C.A, Zak, K, Kolonko, M, Sattler, M, Popowicz, G. | | 登録日 | 2019-03-28 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
7A6S
 
 | | Crystal Structure of Asn173Ser variant of Human Deoxyhypusine Synthase | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Deoxyhypusine synthase, ... | | 著者 | Wator, E, Wilk, P, Grudnik, P. | | 登録日 | 2020-08-26 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Cryo-EM structure of human eIF5A-DHS complex reveals the molecular basis of hypusination-associated neurodegenerative disorders.
Nat Commun, 14, 2023
|
|
