1MGT
 
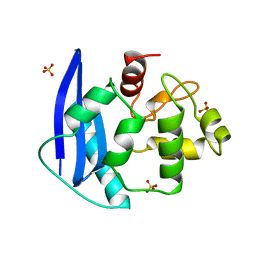 | | CRYSTAL STRUCTURE OF O6-METHYLGUANINE-DNA METHYLTRANSFERASE FROM HYPERTHERMOPHILIC ARCHAEON PYROCOCCUS KODAKARAENSIS STRAIN KOD1 | | 分子名称: | PROTEIN (O6-METHYLGUANINE-DNA METHYLTRANSFERASE), SULFATE ION | | 著者 | Hashimoto, H, Inoue, T, Nishioka, M, Fujiwara, S, Takagi, M, Imanaka, T, Kai, Y. | | 登録日 | 1999-01-12 | | 公開日 | 2000-01-07 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Hyperthermostable protein structure maintained by intra and inter-helix ion-pairs in archaeal O6-methylguanine-DNA methyltransferase.
J.Mol.Biol., 292, 1999
|
|
4FNC
 
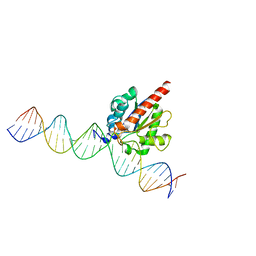 | | Human TDG in a post-reactive complex with 5-hydroxymethyluracil (5hmU) | | 分子名称: | 5-HYDROXYMETHYL URACIL, DNA (28-MER), DNA (29-MER), ... | | 著者 | Hashimoto, H, Hong, S, Bhagwat, A.S, Zhang, X, Cheng, X. | | 登録日 | 2012-06-19 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.493 Å) | | 主引用文献 | Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: its structural basis and implications for active DNA demethylation.
Nucleic Acids Res., 40, 2012
|
|
6DNZ
 
 | | Trypanosoma brucei PRMT1 enzyme-prozyme heterotetrameric complex with AdoHcy | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Arginine N-methyltransferase, putative, ... | | 著者 | Hashimoto, H, Kafkova, L, Jordan, K, Read, L.K, Debler, E.W. | | 登録日 | 2018-06-08 | | 公開日 | 2019-06-12 | | 最終更新日 | 2020-02-12 | | 実験手法 | X-RAY DIFFRACTION (2.384 Å) | | 主引用文献 | Structural Basis of Protein Arginine Methyltransferase Activation by a Catalytically Dead Homolog (Prozyme).
J.Mol.Biol., 432, 2020
|
|
5CZZ
 
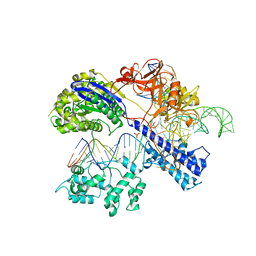 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGAAT PAM) | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | 著者 | Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2015-08-01 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
5K5L
 
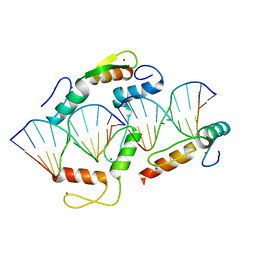 | |
5K5H
 
 | |
5KE8
 
 | |
5K5J
 
 | |
5KE9
 
 | |
5KEA
 
 | |
5KL6
 
 | | Wilms Tumor Protein (WT1) Q369R ZnF2-4 in complex with DNA | | 分子名称: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL4
 
 | |
5KKQ
 
 | | Homo sapiens CCCTC-binding factor (CTCF) ZnF3-7 and DNA complex structure | | 分子名称: | DNA (5'-D(*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*GP*GP*CP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*GP*CP*CP*CP*CP*CP*TP*GP*CP*TP*GP*GP*C)-3'), Transcriptional repressor CTCF, ... | | 著者 | Hashimoto, H, Wang, D, Cheng, X. | | 登録日 | 2016-06-22 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.744 Å) | | 主引用文献 | Structural Basis for the Versatile and Methylation-Dependent Binding of CTCF to DNA.
Mol. Cell, 66, 2017
|
|
5KL5
 
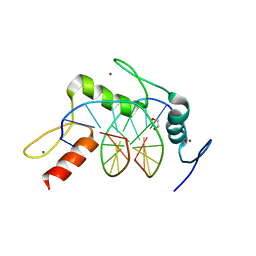 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with carboxylated DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.289 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL2
 
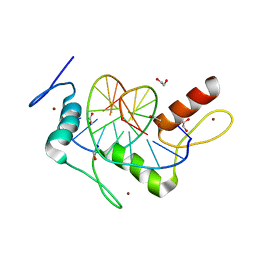 | | Wilms Tumor Protein (WT1) ZnF2-4 in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.692 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL3
 
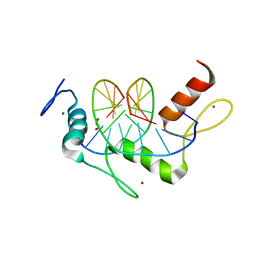 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | 著者 | Hashimoto, H, Cheng, X. | | 登録日 | 2016-06-23 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.449 Å) | | 主引用文献 | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
8XI6
 
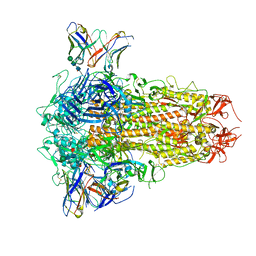 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | 登録日 | 2023-12-19 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 98, 2024
|
|
1O5P
 
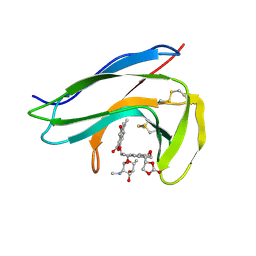 | | Solution Structure of holo-Neocarzinostatin | | 分子名称: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | 著者 | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | 登録日 | 2003-10-04 | | 公開日 | 2003-10-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
7RDN
 
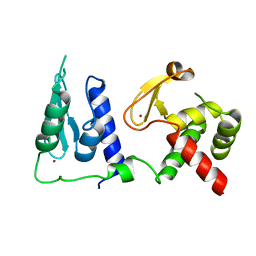 | | Crystal structure of S. cerevisiae pre-mRNA leakage protein 39 (Pml39) | | 分子名称: | Pre-mRNA leakage protein 39, ZINC ION | | 著者 | Hashimoto, H, Ramirez, D.H, Pawlak, N, Blobel, G, Palancade, B, Debler, E.W. | | 登録日 | 2021-07-09 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules.
Sci Rep, 12, 2022
|
|
1EHF
 
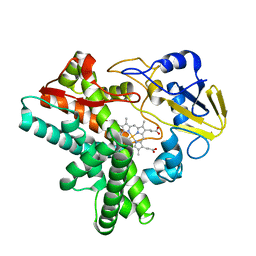 | |
1EHE
 
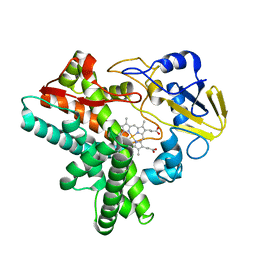 | |
1JFB
 
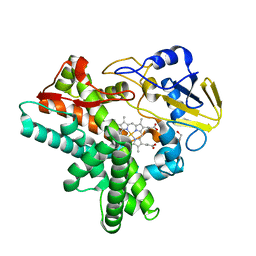 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferric resting state at atomic resolution | | 分子名称: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, nitric-oxide reductase cytochrome P450 55A1 | | 著者 | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2001-06-20 | | 公開日 | 2001-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1EHG
 
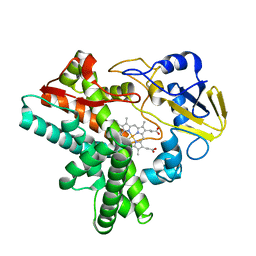 | |
1JFC
 
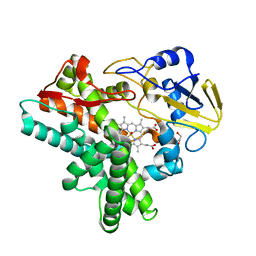 | | X-ray structure of nitric oxide reductase (cytochrome P450nor) in the ferrous CO state at atomic resolution | | 分子名称: | CARBON MONOXIDE, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Shimizu, H, Adachi, S, Park, S.Y, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2001-06-20 | | 公開日 | 2001-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | X-ray structure of nitric oxide reductase (cytochrome P450nor) at atomic resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4GEL
 
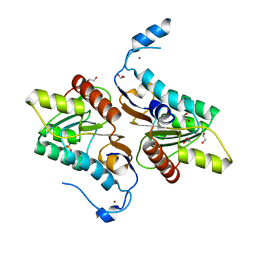 | | Crystal structure of Zucchini | | 分子名称: | 1,2-ETHANEDIOL, Mitochondrial cardiolipin hydrolase, PHOSPHATE ION, ... | | 著者 | Nishimasu, H, Fukuhara, S, Ishitani, R, Nureki, O. | | 登録日 | 2012-08-02 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.756 Å) | | 主引用文献 | Structure and function of Zucchini endoribonuclease in piRNA biogenesis
Nature, 491, 2012
|
|
