2ZIH
 
 | |
2ZII
 
 | |
4U0G
 
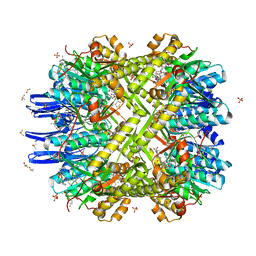 | | Crystal Structure of M. tuberculosis ClpP1P2 bound to ADEP and agonist | | 分子名称: | ADEP-2B5Me, ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2, ... | | 著者 | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | 登録日 | 2014-07-11 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.1978 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U0H
 
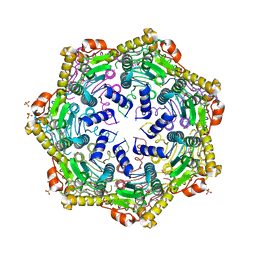 | | Crystal Structure of M. tuberculosis ClpP1P1 | | 分子名称: | ATP-dependent Clp protease proteolytic subunit 1, SULFATE ION | | 著者 | Schmitz, K.R, Carney, D.W, Sello, J.K, Sauer, R.T. | | 登録日 | 2014-07-11 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.2479 Å) | | 主引用文献 | Crystal structure of Mycobacterium tuberculosis ClpP1P2 suggests a model for peptidase activation by AAA+ partner binding and substrate delivery.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3KN1
 
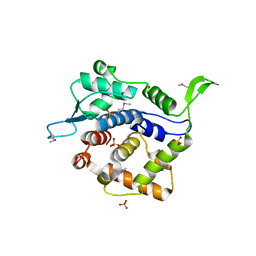 | | Crystal Structure of Golgi Phosphoprotein 3 N-term Truncation Variant | | 分子名称: | Golgi phosphoprotein 3, SULFATE ION | | 著者 | Schmitz, K.R, Bessman, N.J, Setty, T.G, Ferguson, K.M. | | 登録日 | 2009-11-11 | | 公開日 | 2009-12-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | PtdIns4P recognition by Vps74/GOLPH3 links PtdIns 4-kinase signaling to retrograde Golgi trafficking.
J.Cell Biol., 187, 2009
|
|
4KRL
 
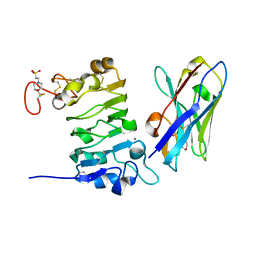 | |
4KRM
 
 | |
4KRP
 
 | |
4KRO
 
 | |
4KRN
 
 | | Nanobody/VHH domain EgA1 | | 分子名称: | Nanobody/VHH domain EgA1, SULFATE ION | | 著者 | Ferguson, K.M, Schmitz, K.R. | | 登録日 | 2013-05-16 | | 公開日 | 2013-08-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.553 Å) | | 主引用文献 | Structural Evaluation of EGFR Inhibition Mechanisms for Nanobodies/VHH Domains.
Structure, 21, 2013
|
|
3EGA
 
 | |
3EGB
 
 | |
1YY8
 
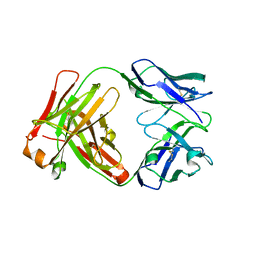 | | Crystal structure of the Fab fragment from the monoclonal antibody cetuximab/Erbitux/IMC-C225 | | 分子名称: | Cetuximab Fab Heavy chain, Cetuximab Fab Light chain | | 著者 | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | 登録日 | 2005-02-24 | | 公開日 | 2005-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
1YY9
 
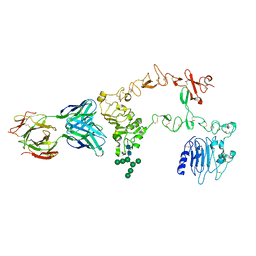 | | Structure of the extracellular domain of the epidermal growth factor receptor in complex with the Fab fragment of cetuximab/Erbitux/IMC-C225 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cetuximab Fab Heavy chain, ... | | 著者 | Li, S, Schmitz, K.R, Jeffrey, P.D, Wiltzius, J.J.W, Kussie, P, Ferguson, K.M. | | 登録日 | 2005-02-24 | | 公開日 | 2005-04-26 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.605 Å) | | 主引用文献 | Structural basis for inhibition of the epidermal growth factor receptor by cetuximab
Cancer Cell, 7, 2005
|
|
4I5O
 
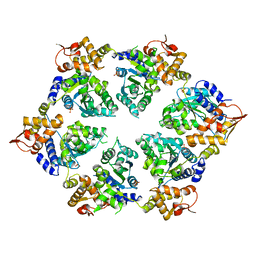 | | Crystal Structure of W-W-R ClpX Hexamer | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | 著者 | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | 登録日 | 2012-11-28 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (4.4787 Å) | | 主引用文献 | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I9K
 
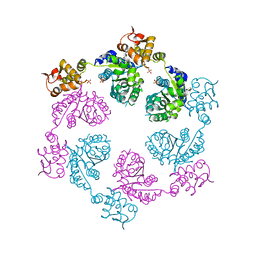 | | Crystal structure of symmetric W-W-W ClpX Hexamer | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | 著者 | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | 登録日 | 2012-12-05 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5.0003 Å) | | 主引用文献 | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I63
 
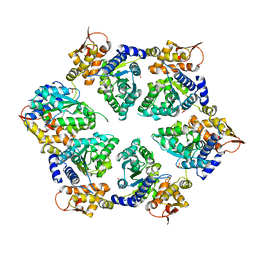 | | Crystal Structure of E-R ClpX Hexamer | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | 著者 | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | 登録日 | 2012-11-29 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5.709 Å) | | 主引用文献 | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I34
 
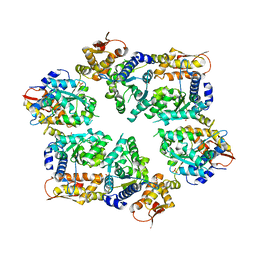 | | Crystal Structure of W-W-W ClpX Hexamer | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | 著者 | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | 登録日 | 2012-11-23 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (4.1218 Å) | | 主引用文献 | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I81
 
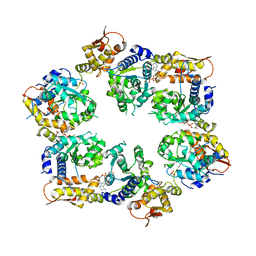 | | Crystal Structure of ATPgS bound ClpX Hexamer | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit ClpX, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, SULFATE ION | | 著者 | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | 登録日 | 2012-12-01 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.8182 Å) | | 主引用文献 | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4I4L
 
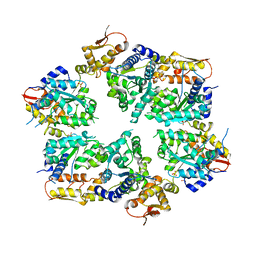 | | Crystal Structure of Nucleotide-Bound W-W-W ClpX Hexamer | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, SULFATE ION | | 著者 | Glynn, S.E, Nager, A.R, Stinson, B.S, Schmitz, K.R, Baker, T.A, Sauer, R.T. | | 登録日 | 2012-11-27 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.6981 Å) | | 主引用文献 | Nucleotide Binding and Conformational Switching in the Hexameric Ring of a AAA+ Machine.
Cell(Cambridge,Mass.), 153, 2013
|
|
4WPC
 
 | |
3OSE
 
 | |
3OST
 
 | |
3OSM
 
 | |
5JI2
 
 | | HslU L199Q in HslUV complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | 著者 | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | 登録日 | 2016-04-21 | | 公開日 | 2016-11-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.307 Å) | | 主引用文献 | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
