5MNS
 
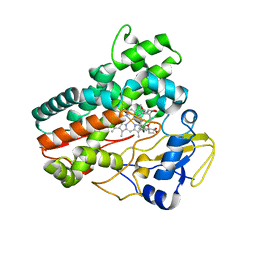 | | Structural and functional characterization of OleP in complex with 6DEB in sodium formate | | 分子名称: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Parisi, G, Savino, C, Montemiglio, L.C, Vallone, B. | | 登録日 | 2016-12-13 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Substrate-induced conformational change in cytochrome P450 OleP.
FASEB J., 33, 2019
|
|
5MNV
 
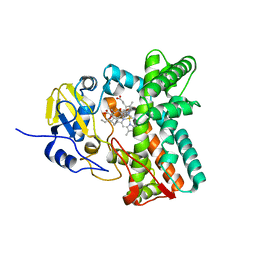 | |
6ZHZ
 
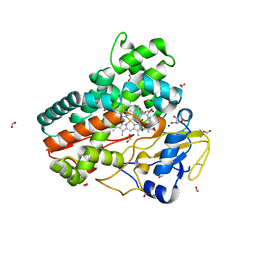 | | OleP-oleandolide(DEO) in high salt crystallization conditions | | 分子名称: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P-450, ... | | 著者 | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Cecchetti, C. | | 登録日 | 2020-06-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZI2
 
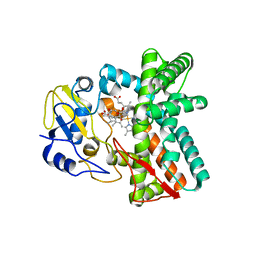 | | OleP-oleandolide(DEO) in low salt crystallization conditions | | 分子名称: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Savino, C, Montemiglio, L.C, Vallone, B, Parisi, G, Cecchetti, C. | | 登録日 | 2020-06-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.93 Å) | | 主引用文献 | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZI7
 
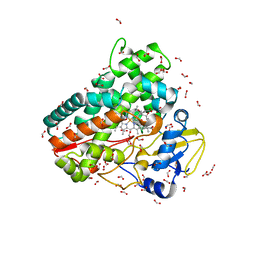 | | Crystal structure of OleP-oleandolide(DEO) bound to L-rhamnose | | 分子名称: | (3~{R},4~{S},5~{R},6~{S},7~{S},9~{S},11~{R},12~{S},13~{R},14~{R})-3,5,7,9,11,13,14-heptamethyl-4,6,12-tris(oxidanyl)-1-oxacyclotetradecane-2,10-dione, Cytochrome P-450, FORMIC ACID, ... | | 著者 | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | 登録日 | 2020-06-25 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
6ZI3
 
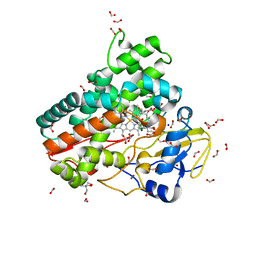 | | Crystal structure of OleP-6DEB bound to L-rhamnose | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, ... | | 著者 | Montemiglio, L.C, Savino, C, Vallone, B, Parisi, G, Freda, I. | | 登録日 | 2020-06-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Dissecting the Cytochrome P450 OleP Substrate Specificity: Evidence for a Preferential Substrate.
Biomolecules, 10, 2020
|
|
7MJS
 
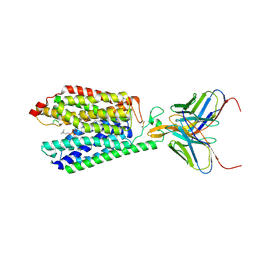 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | 著者 | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | 登録日 | 2021-04-20 | | 公開日 | 2021-06-16 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
4XE3
 
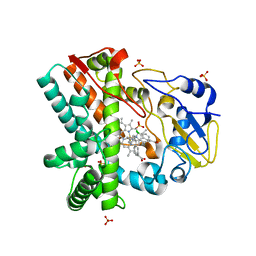 | | OleP, the cytochrome P450 epoxidase from Streptomyces antibioticus involved in Oleandomycin biosynthesis: functional analysis and crystallographic structure in complex with clotrimazole. | | 分子名称: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Montemiglio, L.C, Parisi, G, Scaglione, A, Savino, C, Vallone, B. | | 登録日 | 2014-12-22 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Functional analysis and crystallographic structure of clotrimazole bound OleP, a cytochrome P450 epoxidase from Streptomyces antibioticus involved in oleandomycin biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
7KC4
 
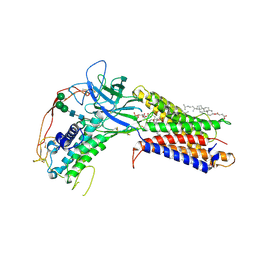 | | Human WLS in complex with WNT8A | | 分子名称: | 1-CIS-9-OCTADECANOYL-2-CIS-9-HEXADECANOYL PHOSPHATIDYL GLYCEROL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Nygaard, R, Jia, Y, Kim, J, Ross, D, Parisi, G, Clarke, O.B, Virshup, D.M, Mancia, F. | | 登録日 | 2020-10-05 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Structural Basis of WLS/Evi-Mediated Wnt Transport and Secretion.
Cell, 184, 2021
|
|
6H6J
 
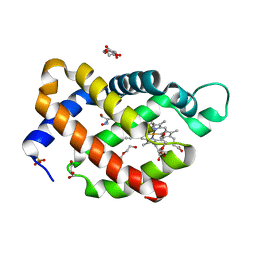 | | Carbomonoxy murine neuroglobin Gly-loop mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBON MONOXIDE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | 登録日 | 2018-07-27 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H5Z
 
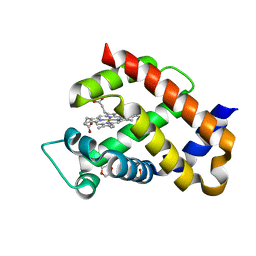 | | Ferric murine neuroglobin F106A mutant | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | 登録日 | 2018-07-25 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H6I
 
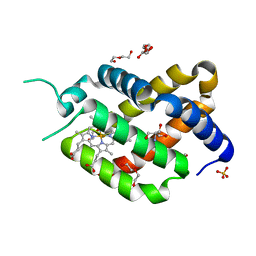 | | Ferric murine neuroglobin Gly-loop mutant | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | 登録日 | 2018-07-27 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H6C
 
 | | Carbomonoxy murine neuroglobin F106A mutant | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, CARBON MONOXIDE, GLYCEROL, ... | | 著者 | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | 登録日 | 2018-07-27 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
5N9X
 
 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, ligand bound structure | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Adenylation domain, MAGNESIUM ION, ... | | 著者 | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | 登録日 | 2017-02-27 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
5N9W
 
 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, apo structure | | 分子名称: | ACETATE ION, Adenylation domain | | 著者 | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | 登録日 | 2017-02-27 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.456 Å) | | 主引用文献 | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
7NMN
 
 | | Rabbit HCN4 stabilised in amphipol A8-35 | | 分子名称: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Rabbit HCN4 | | 著者 | Chaves-Sanjuan, A. | | 登録日 | 2021-02-23 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP4
 
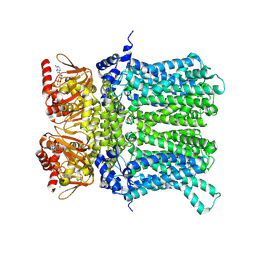 | | cAMP-bound rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Giese, H, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | 登録日 | 2021-02-26 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7NP3
 
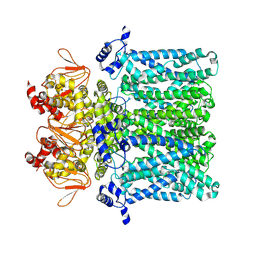 | | cAMP-free rabbit HCN4 stabilized in LMNG-CHS detergent mixture | | 分子名称: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | 著者 | Giese, H.M, Chaves-Sanjuan, A, Saponaro, A, Clarke, O, Bolognesi, M, Mancia, F, Hendrickson, W.A, Thiel, G, Santoro, B, Moroni, A. | | 登録日 | 2021-02-26 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Gating movements and ion permeation in HCN4 pacemaker channels.
Mol.Cell, 81, 2021
|
|
7OHD
 
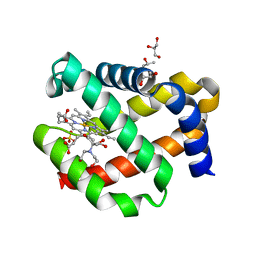 | | CRYSTAL STRUCTURE OF FERRIC MURINE NEUROGLOBIN CDLESS MUTANT | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ISOPROPYL ALCOHOL, ... | | 著者 | Exertier, C, Freda, I, Montemiglio, L.C, Savino, C, Cerutti, G, Gugole, E, Vallone, B. | | 登録日 | 2021-05-10 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Probing the Role of Murine Neuroglobin CDloop-D-Helix Unit in CO Ligand Binding and Structural Dynamics.
Acs Chem.Biol., 17, 2022
|
|
7Q6X
 
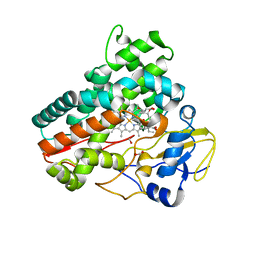 | | OleP mutant S240Y in complex with 6DEB | | 分子名称: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | 著者 | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | 登録日 | 2021-11-09 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
7Q89
 
 | | OleP mutant G92W in complex with 6DEB | | 分子名称: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | 著者 | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | 登録日 | 2021-11-10 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
7Q6R
 
 | | OleP mutant E89Y in complex with 6DEB | | 分子名称: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, FORMIC ACID, ... | | 著者 | Savino, C, Montemiglio, L.C, Vallone, B, Exertier, C, Freda, I, Gugole, E. | | 登録日 | 2021-11-09 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Point Mutations at a Key Site Alter the Cytochrome P450 OleP Structural Dynamics.
Biomolecules, 12, 2021
|
|
