1CDQ
 
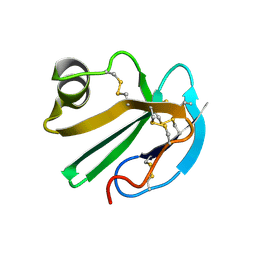 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | 分子名称: | CD59 | | 著者 | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | 登録日 | 1994-06-01 | | 公開日 | 1994-09-30 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
1CDR
 
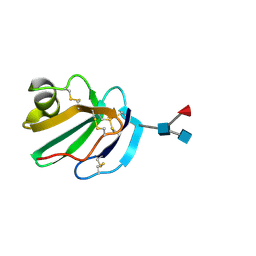 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | 著者 | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | 登録日 | 1994-06-01 | | 公開日 | 1994-09-30 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
1JCS
 
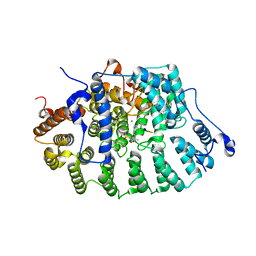 | | CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH THE PEPTIDE SUBSTRATE TKCVFM AND AN ANALOG OF FARNESYL DIPHOSPHATE | | 分子名称: | ACETIC ACID, PROTEIN FARNESYLTRANSFERASE, ALPHA SUBUNIT, ... | | 著者 | Long, S.B, Casey, P.J, Beese, L.S. | | 登録日 | 2001-06-11 | | 公開日 | 2001-11-02 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
6JXM
 
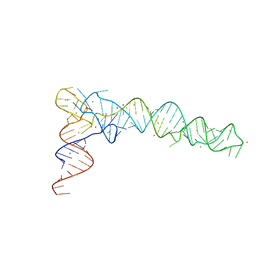 | | Crystal Structure of phi29 pRNA domain II | | 分子名称: | BARIUM ION, MAGNESIUM ION, RNA (97-mer) | | 著者 | Cai, R, Price, I.R, Ding, F, Wu, F, Chen, T, Zhang, Y, Liu, G, Jardine, P.J, Lu, C, Ke, A. | | 登録日 | 2019-04-24 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | ATP/ADP modulates gp16-pRNA conformational change in the Phi29 DNA packaging motor.
Nucleic Acids Res., 47, 2019
|
|
2V3V
 
 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | 分子名称: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | 著者 | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J, Brondino, C.D, Romao, M.J. | | 登録日 | 2007-06-22 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
1BSX
 
 | | STRUCTURE AND SPECIFICITY OF NUCLEAR RECEPTOR-COACTIVATOR INTERACTIONS | | 分子名称: | 3,5,3'TRIIODOTHYRONINE, PROTEIN (GRIP1), PROTEIN (THYROID HORMONE RECEPTOR BETA) | | 著者 | Wagner, R.L, Darimont, B.D, Apriletti, J.W, Stallcup, M.R, Kushner, P.J, Baxter, J.D, Fletterick, R.J, Yamamoto, K.R. | | 登録日 | 1998-08-31 | | 公開日 | 1999-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structure and specificity of nuclear receptor-coactivator interactions.
Genes Dev., 12, 1998
|
|
1BWR
 
 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | 分子名称: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | 著者 | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | 登録日 | 1998-09-27 | | 公開日 | 1999-05-18 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
1C5F
 
 | | CRYSTAL STRUCTURE OF THE CYCLOPHILIN-LIKE DOMAIN FROM BRUGIA MALAYI COMPLEXED WITH CYCLOSPORIN A | | 分子名称: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 1 | | 著者 | Ellis, P.J, Carlow, C.K.S, Ma, D, Kuhn, P. | | 登録日 | 1999-11-22 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Crystal Structure of the Complex of Brugia Malayi Cyclophilin and Cyclosporin A.
Biochemistry, 39, 2000
|
|
1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | 分子名称: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | 著者 | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | 登録日 | 2004-02-04 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
3CQP
 
 | | Human SOD1 G85R Variant, Structure I | | 分子名称: | COPPER (II) ION, MALONATE ION, Superoxide dismutase [Cu-Zn], ... | | 著者 | Cao, X, Antonyuk, S, Seetharaman, S.V, Whitson, L.J, Taylor, A.B, Holloway, S.P, Strange, R.W, Doucette, P.A, Valentine, J.S, Tiwari, A, Hayward, L.J, Padua, S, Cohlberg, J.A, Hasnain, S.S, Hart, P.J. | | 登録日 | 2008-04-03 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the G85R Variant of SOD1 in Familial Amyotrophic Lateral Sclerosis.
J.Biol.Chem., 283, 2008
|
|
4U1J
 
 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GAG protein, ... | | 著者 | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | 登録日 | 2014-07-15 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
4U1N
 
 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | 登録日 | 2014-07-15 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
4TVY
 
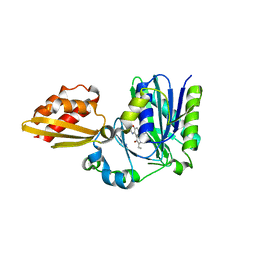 | | Apo resorufin ligase | | 分子名称: | 5-(3,7-dihydroxy-10H-phenoxazin-2-yl)pentanamide, Lipoate-protein ligase A | | 著者 | Goldman, P.J, Drennan, C.L. | | 登録日 | 2014-06-28 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.151 Å) | | 主引用文献 | Computational design of a red fluorophore ligase for site-specific protein labeling in living cells.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4U1S
 
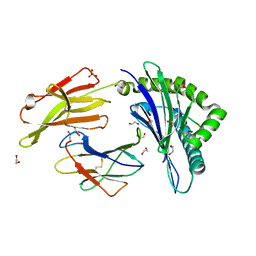 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | 登録日 | 2014-07-16 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
4U3V
 
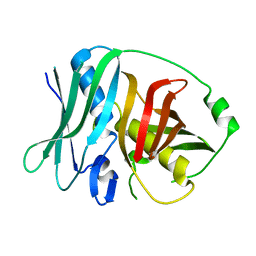 | |
4U1K
 
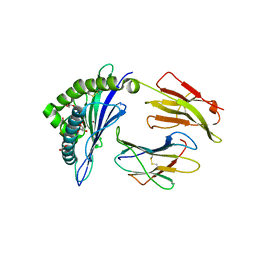 | | HLA class I micropolymorphisms determine peptide-HLA landscape and dictate differential HIV-1 escape through identical epitopes | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | 著者 | Rizkallah, P.J, Cole, D.K, Fuller, A, Sewell, A.K. | | 登録日 | 2014-07-15 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | A molecular switch in immunodominant HIV-1-specific CD8 T-cell epitopes shapes differential HLA-restricted escape.
Retrovirology, 12, 2015
|
|
4U92
 
 | | Crystal structure of a DNA/Ba2+ G-quadruplex containing a water-mediated C-tetrad | | 分子名称: | BARIUM ION, DNA (5'-D(*CP*CP*AP*KP*GP*CP*GP*TP*GP*G)-3'), MAGNESIUM ION | | 著者 | Paukstelis, P.J, Zhang, D, Huang, T, Lukeman, P. | | 登録日 | 2014-08-05 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of a DNA/Ba2+ G-quadruplex containing a water-mediated C-tetrad.
Nucleic Acids Res., 42, 2014
|
|
1CRQ
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | 分子名称: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | 登録日 | 1993-11-24 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CRP
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | 分子名称: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | 登録日 | 1993-11-24 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CRR
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | 分子名称: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | 登録日 | 1993-11-24 | | 公開日 | 1994-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
4K1P
 
 | | Structure of the NheA component of the Nhe toxin from Bacillus cereus | | 分子名称: | 1,2-ETHANEDIOL, NheA, SULFATE ION | | 著者 | Ganash, M, Phung, D, Artymiuk, P.J. | | 登録日 | 2013-04-05 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the NheA Component of the Nhe Toxin from Bacillus cereus: Implications for Function.
Plos One, 8, 2013
|
|
1IJG
 
 | | Structure of the Bacteriophage phi29 Head-Tail Connector Protein | | 分子名称: | UPPER COLLAR PROTEIN | | 著者 | Simpson, A.A, Leiman, P.G, Tao, Y, He, Y, Badasso, M, Jardine, P.J, Anderson, D.L, Rossmann, M.G. | | 登録日 | 2001-04-26 | | 公開日 | 2001-05-09 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure determination of the head-tail connector of bacteriophage phi29.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IEL
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with Ceftazidime | | 分子名称: | ACYLATED CEFTAZIDIME, PHOSPHATE ION, beta-lactamase | | 著者 | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | 登録日 | 2001-04-10 | | 公開日 | 2001-08-15 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1IEM
 
 | | Crystal Structure of AmpC beta-lactamase from E. coli in Complex with a Boronic Acid Inhibitor (1, CefB4) | | 分子名称: | PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, beta-lactamase | | 著者 | Powers, R.A, Caselli, E, Focia, P.J, Prati, F, Shoichet, B.K. | | 登録日 | 2001-04-10 | | 公開日 | 2001-08-15 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structures of ceftazidime and its transition-state analogue in complex with AmpC beta-lactamase: implications for resistance mutations and inhibitor design.
Biochemistry, 40, 2001
|
|
1IHV
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | HIV-1 INTEGRASE | | 著者 | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | 登録日 | 1995-05-12 | | 公開日 | 1996-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
