6K0W
 
 | |
2KZ9
 
 | |
7WVL
 
 | |
6JQN
 
 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and OCoA | | 分子名称: | Bifunctional protein PaaZ, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OCTANOYL-COENZYME A | | 著者 | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQL
 
 | | Structure of PaaZ, a bifunctional enzyme | | 分子名称: | Bifunctional protein PaaZ | | 著者 | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQO
 
 | | Structure of PaaZ, a bifunctional enzyme in complex with NADP+ and CCoA | | 分子名称: | Bifunctional protein PaaZ, CROTONYL COENZYME A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
6JQM
 
 | | Structure of PaaZ with NADPH | | 分子名称: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | 登録日 | 2019-03-31 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
8U7Z
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U81
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U83
 
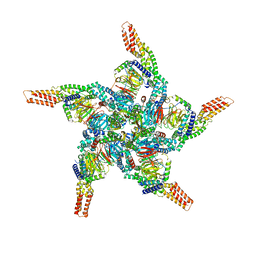 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.975 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U80
 
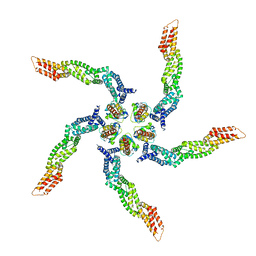 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(BTB)/Cullin3(NTD) | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3 | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5D4S
 
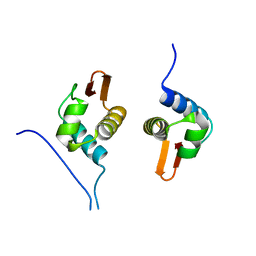 | | Crystal Structure of AraR(DBD) in complex with operator ORX1 | | 分子名称: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*AP*AP*TP*AP*CP*AP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*TP*GP*TP*AP*TP*T)-3') | | 著者 | Jain, D, Narayanan, N, Nair, D.T. | | 登録日 | 2015-08-08 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.972 Å) | | 主引用文献 | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
5D4R
 
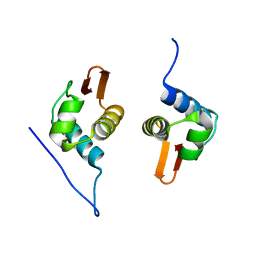 | | Crystal Structure of AraR(DBD) in complex with operator ORE1 | | 分子名称: | Arabinose metabolism transcriptional repressor, DNA (5'-D(*AP*TP*AP*TP*TP*TP*GP*TP*AP*CP*GP*TP*AP*CP*TP*AP*AP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*TP*TP*AP*GP*TP*AP*CP*GP*TP*AP*CP*AP*AP*AP*TP*A)-3') | | 著者 | Jain, D, Narayanan, N, Nair, D.T. | | 登録日 | 2015-08-08 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Plasticity in Repressor-DNA Interactions Neutralizes Loss of Symmetry in Bipartite Operators.
J.Biol.Chem., 291, 2016
|
|
2K88
 
 | | Association of subunit d (Vma6p) and E (Vma4p) with G (Vma10p) and the NMR solution structure of subunit G (G1-59) of the Saccharomyces cerevisiae V1VO ATPase | | 分子名称: | Vacuolar proton pump subunit G | | 著者 | Sankaranarayanan, N, Gayen, S, Thaker, Y, Subramanian, V, Manimekalai, M.S.S, Gruber, G. | | 登録日 | 2008-09-04 | | 公開日 | 2009-08-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Assembly of subunit d (Vma6p) and G (Vma10p) and the NMR solution structure of subunit G (G(1-59)) of the Saccharomyces cerevisiae V(1)V(O) ATPase.
Biochim.Biophys.Acta, 1787, 2009
|
|
4IRD
 
 | | Structure of Polymerase-DNA complex | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | 著者 | Nair, D.T, Sharma, A. | | 登録日 | 2013-01-14 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IR1
 
 | | Polymerase-DNA Complex | | 分子名称: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | 著者 | Sharma, A, Nair, D.T. | | 登録日 | 2013-01-14 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IRC
 
 | | Polymerase-DNA complex | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | 著者 | Nair, D.T, Sharma, A. | | 登録日 | 2013-01-14 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IR9
 
 | | Polymerase-DNA complex | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(P*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | 著者 | Sharma, A, Nair, D.T. | | 登録日 | 2013-01-14 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IRK
 
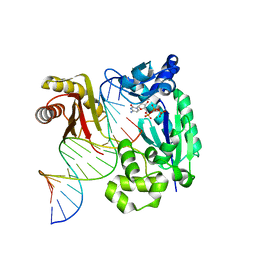 | | structure of Polymerase-DNA complex, dna | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*A*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*(DOC))-3'), DNA (5'-D(*TP*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | 著者 | Nair, D.T, Sharma, A. | | 登録日 | 2013-01-14 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4Q44
 
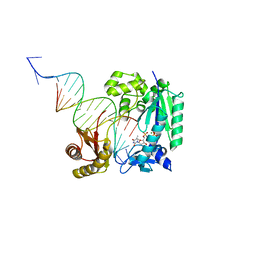 | | Polymerase-Damaged DNA Complex | | 分子名称: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*TP*CP*TP*AP*(RDG)P*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA polymerase IV, ... | | 著者 | Nair, D.T, Kottur, J, Sharma, A. | | 登録日 | 2014-04-13 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
4Q43
 
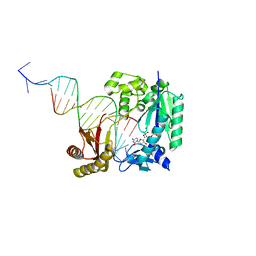 | | Polymerase-damaged DNA complex | | 分子名称: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*T*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*CP*TP*(RDG)P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | 著者 | Kottur, J, Sharma, A, Nair, D.T. | | 登録日 | 2014-04-13 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
4Q45
 
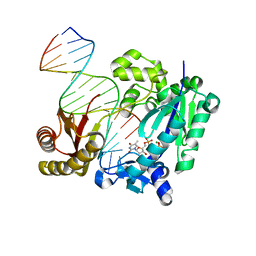 | | DNA Polymerase- damaged DNA complex | | 分子名称: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*TP*CP*TP*A*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*CP*TP*AP*GP*GP*(RDG)P*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | 著者 | Kottur, J, Sharma, A, Nair, D.T. | | 登録日 | 2014-04-13 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.176 Å) | | 主引用文献 | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
5EPQ
 
 | | Structure at 1.75 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Banerjee, S, Chavas, L.M.G, Ramaswamy, S. | | 登録日 | 2015-11-12 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
