3T4R
 
 | |
4ABM
 
 | | Crystal Structure of CHMP4B hairpin | | 分子名称: | CHARGED MULTIVESICULAR BODY PROTEIN 4B | | 著者 | Martinelli, N, Hartlieb, B, Usami, Y, Sabin, C, Dordor, A, Ribeiro, E.A, Gottlinger, H, Weissenhorn, W. | | 登録日 | 2011-12-08 | | 公開日 | 2012-05-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Cc2D1A is a Regulator of Escrt-III Chmp4B.
J.Mol.Biol., 419, 2012
|
|
7LBE
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with neutralizing fabs 13H11 and MSL-109 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | 著者 | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | 登録日 | 2021-01-07 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBG
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Transforming growth factor beta receptor type 3 and neutralizing fabs 13H11 and MSL-109 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | 著者 | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | 登録日 | 2021-01-07 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7LBF
 
 | | CryoEM structure of the HCMV Trimer gHgLgO in complex with human Platelet-derived growth factor receptor alpha and neutralizing fabs 13H11 and MSL-109 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | 著者 | Kschonsak, M, Rouge, L, Arthur, C.P, Hoangdung, H, Patel, N, Kim, I, Johnson, M, Kraft, E, Rohou, A.L, Gill, A, Martinez-Martin, N, Payandeh, J, Ciferri, C. | | 登録日 | 2021-01-07 | | 公開日 | 2021-03-10 | | 最終更新日 | 2021-03-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structures of HCMV Trimer reveal the basis for receptor recognition and cell entry.
Cell, 184, 2021
|
|
7T4S
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with NRP2 and neutralizing fabs 8I21 and 13H11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein H, ... | | 著者 | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | 登録日 | 2021-12-10 | | 公開日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4Q
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with neutralizing fabs 2C12, 7I13 and 13H11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | 著者 | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | 登録日 | 2021-12-10 | | 公開日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
7T4R
 
 | | CryoEM structure of the HCMV Pentamer gH/gL/UL128/UL130/UL131A in complex with THBD and neutralizing fabs MSL-109 and 13H11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | 著者 | Kschonsak, M, Johnson, M.C, Schelling, R, Green, E.M, Rouge, L, Ho, H, Patel, N, Kilic, C, Kraft, E, Arthur, C.P, Rohou, A.L, Comps-Agrar, L, Martinez-Martin, N, Perez, L, Payandeh, J, Ciferri, C. | | 登録日 | 2021-12-10 | | 公開日 | 2022-03-23 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for HCMV Pentamer receptor recognition and antibody neutralization.
Sci Adv, 8, 2022
|
|
6EI6
 
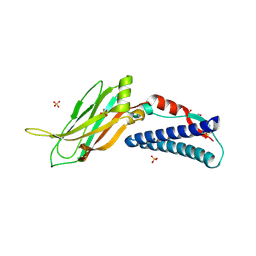 | | CC2D1B coordinates ESRCT-III activity during the mitotic reformation of the nuclear envelope | | 分子名称: | Coiled-coil and C2 domain-containing protein 1-like, DI(HYDROXYETHYL)ETHER, SULFATE ION | | 著者 | Ventimiglia, L.N, Cuesta-Geijo, M.A, Martinelli, N, Caballe, A, Macheboeuf, P, Miguet, N, Parnham, I.M, Olmos, Y, Carlton, J.G, Weissehorn, W, martin-Serrano, J. | | 登録日 | 2017-09-18 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.461 Å) | | 主引用文献 | CC2D1B Coordinates ESCRT-III Activity during the Mitotic Reformation of the Nuclear Envelope.
Dev. Cell, 47, 2018
|
|
5I8L
 
 | | Crystal structure of the full-length cell wall-binding module of Cpl7 mutant R223A | | 分子名称: | GLYCEROL, Lysozyme | | 著者 | Bernardo-Garcia, N, Silva-Martin, N, Uson, I, Hermoso, J.A. | | 登録日 | 2016-02-19 | | 公開日 | 2017-03-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Deciphering how Cpl-7 cell wall-binding repeats recognize the bacterial peptidoglycan.
Sci Rep, 7, 2017
|
|
3H3X
 
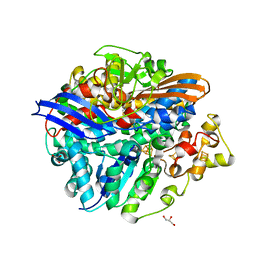 | | Structure of the V74M large subunit mutant of NI-FE hydrogenase in an oxidized state | | 分子名称: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | 著者 | Volbeda, A, Martinez, N, Martin, L, Fontecilla-Camps, J.C. | | 登録日 | 2009-04-17 | | 公開日 | 2009-07-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant
J.Am.Chem.Soc., 131, 2009
|
|
2Y2C
 
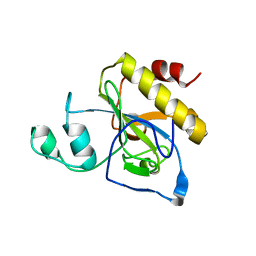 | | crystal structure of AmpD Apoenzyme | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y28
 
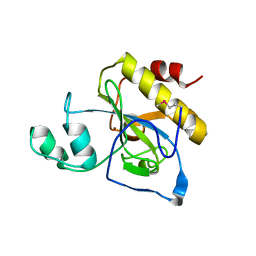 | | crystal structure of Se-Met AmpD derivative | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2B
 
 | | crystal structure of AmpD in complex with reaction products | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, ... | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2E
 
 | | crystal structure of AmpD grown at pH 5.5 | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
2Y2D
 
 | | crystal structure of AmpD holoenzyme | | 分子名称: | 1,6-ANHYDRO-N-ACETYLMURAMYL-L-ALANINE AMIDASE AMPD, ZINC ION | | 著者 | Carrasco-Lopez, C, Rojas-Altuve, A, Zhang, W, Hesek, D, Lee, M, Barbe, S, Andre, I, Silva-Martin, N, Martinez-Ripoll, M, Mobashery, S, Hermoso, J.A. | | 登録日 | 2010-12-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Bacterial Peptidoglycan Amidase Ampd and an Unprecedented Activation Mechanism.
J.Biol.Chem., 286, 2011
|
|
5FM6
 
 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(apo) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, RVB1, ... | | 著者 | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | 登録日 | 2015-11-02 | | 公開日 | 2016-01-20 | | 実験手法 | X-RAY DIFFRACTION (2.997 Å) | | 主引用文献 | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
5FM7
 
 | | Double-heterohexameric rings of full-length Rvb1(ADP)Rvb2(ADP) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, RVB1, RVB2 | | 著者 | Silva-Martin, N, Dauden, M.I, Glatt, S, Hoffmann, N.A, Mueller, C.W. | | 登録日 | 2015-11-02 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.901 Å) | | 主引用文献 | The Combination of X-Ray Crystallography and Cryo-Electron Microscopy Provides Insight Into the Overall Architecture of the Dodecameric Rvb1/Rvb2 Complex.
Plos One, 11, 2016
|
|
7TPR
 
 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | 分子名称: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | 著者 | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | 登録日 | 2022-01-25 | | 公開日 | 2022-04-20 | | 実験手法 | ELECTRON MICROSCOPY (2.39 Å) | | 主引用文献 | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
2X8P
 
 | |
2X8O
 
 | |
3ZHG
 
 | |
4CVD
 
 | |
4CAJ
 
 | | Crystallographic structure of the mouse SIGN-R1 CRD domain in complex with sialic acid | | 分子名称: | CALCIUM ION, CD209 ANTIGEN-LIKE PROTEIN B, CHLORIDE ION, ... | | 著者 | Silva-Martin, N, Bartual, S.G, Hermoso, J.A. | | 登録日 | 2013-10-08 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.191 Å) | | 主引用文献 | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
4CDH
 
 | | Crystallographic structure of the Human Igg1 alpha 2-6 sialilated Fc-Fragment | | 分子名称: | IG GAMMA-1 CHAIN C REGION, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Silva-Martin, N, Bartual, S.G, Hermoso, J.A. | | 登録日 | 2013-10-31 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
