8BBW
 
 | | Crystal structure of medical leech destabilase (low salt) | | 分子名称: | CHLORIDE ION, GLYCEROL, Lysozyme | | 著者 | Marin, E, Bukhdruker, S, Manuvera, V, Kornilov, D, Zinovev, E, Bobrovsky, P, Lazarev, V, Borshchevskiy, V. | | 登録日 | 2022-10-14 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural basis for inhibition of thrombolytic destabilase
from medical leech by physiological sodium concentrations
To Be Published
|
|
8BBU
 
 | | Crystal structure of medical leech destabilase (high salt) | | 分子名称: | GLYCEROL, Lysozyme, MALONATE ION, ... | | 著者 | Marin, E, Bukhdruker, S, Manuvera, V, Kornilov, D, Zinovev, E, Bobrovsky, P, Lazarev, V, Borshchevskiy, V. | | 登録日 | 2022-10-14 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural insights into thrombolytic activity of destabilase from medicinal leech.
Sci Rep, 13, 2023
|
|
4YGI
 
 | |
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | 分子名称: | De novo beta protein | | 著者 | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | 登録日 | 2018-07-19 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2QXU
 
 | |
2QZ2
 
 | | Crystal structure of a glycoside hydrolase family 11 xylanase from Aspergillus niger in complex with xylopentaose | | 分子名称: | Endo-1,4-beta-xylanase I, SODIUM ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ... | | 著者 | Vandermarliere, E, Rombouts, S, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | 登録日 | 2007-08-16 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2Z79
 
 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | 分子名称: | Endo-1,4-beta-xylanase A, GLYCEROL | | 著者 | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | 登録日 | 2007-08-16 | | 公開日 | 2007-12-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
3C7O
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with cellotetraose. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-08 | | 公開日 | 2008-11-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7F
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from bacillus subtilis in complex with xylotriose. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-07 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7H
 
 | | Crystal structure of glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with AXOS-4-0.5. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-07 | | 公開日 | 2008-11-18 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7G
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis in complex with xylotetraose. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, GLYCEROL, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-07 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
3C7E
 
 | | Crystal structure of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase from Bacillus subtilis. | | 分子名称: | CALCIUM ION, Endo-1,4-beta-xylanase, FORMIC ACID, ... | | 著者 | Vandermarliere, E, Bourgois, T.M, Winn, M.D, Van Campenhout, S, Volckaert, G, Strelkov, S.V, Delcour, J.A, Rabijns, A, Courtin, C.M. | | 登録日 | 2008-02-07 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 43 arabinoxylan arabinofuranohydrolase in complex with xylotetraose reveals a different binding mechanism compared with other members of the same family.
Biochem.J., 418, 2009
|
|
2HNX
 
 | | Crystal Structure of aP2 | | 分子名称: | ACETIC ACID, Fatty acid-binding protein, adipocyte, ... | | 著者 | Marr, E, Tardie, M, Carty, M, Brown Phillips, T, Qiu, X, Karam, G. | | 登録日 | 2006-07-13 | | 公開日 | 2006-11-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Expression, purification, crystallization and structure of human adipocyte lipid-binding protein (aP2).
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3SOW
 
 | |
3G7M
 
 | | Structure of the thaumatin-like xylanase inhibitor TLXI | | 分子名称: | GLYCEROL, SODIUM ION, Xylanase inhibitor TL-XI | | 著者 | Vandermarliere, E, Courtin, C.M, Lammens, W, Schoepe, J, Strelkov, S.V. | | 登録日 | 2009-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Crystal structure of the noncompetitive xylanase inhibitor TLXI, member of the small thaumatin-like protein family.
Proteins, 78, 2010
|
|
3SOU
 
 | |
3SOX
 
 | |
2QZ3
 
 | | Crystal structure of a glycoside hydrolase family 11 xylanase from Bacillus subtilis in complex with xylotetraose | | 分子名称: | ACETIC ACID, Endo-1,4-beta-xylanase A, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | 登録日 | 2007-08-16 | | 公開日 | 2007-12-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
2QXT
 
 | |
5L33
 
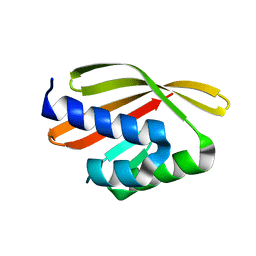 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | denovo NTF2 | | 著者 | Oberdorfer, G, Marcos, E, Basanta, B, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-08-03 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
1T2N
 
 | |
1T4M
 
 | |
5TPJ
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | denovo NTF2 | | 著者 | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-10-20 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.101 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TPH
 
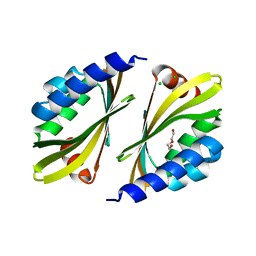 | | Crystal structure of a de novo designed protein homodimer with curved beta-sheet | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, de novo NTF2 homodimer | | 著者 | Basanta, B, Marcos, E, Oberdorfer, G, Chidyausiku, T.M, Sankaran, B, Baker, D. | | 登録日 | 2016-10-20 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | 分子名称: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | 著者 | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | 登録日 | 2016-10-27 | | 公開日 | 2017-01-25 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (3.005 Å) | | 主引用文献 | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
