1UD7
 
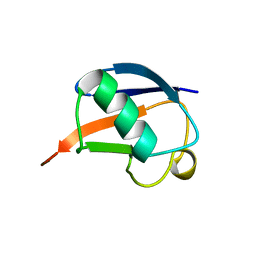 | | SOLUTION STRUCTURE OF THE DESIGNED HYDROPHOBIC CORE MUTANT OF UBIQUITIN, 1D7 | | 分子名称: | PROTEIN (UBIQUITIN CORE MUTANT 1D7) | | 著者 | Johnson, E.C, Lazar, G.A, Desjarlais, J.R, Handel, T.M. | | 登録日 | 1999-04-07 | | 公開日 | 1999-05-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of a designed hydrophobic core variant of ubiquitin.
Structure Fold.Des., 7, 1999
|
|
2J9K
 
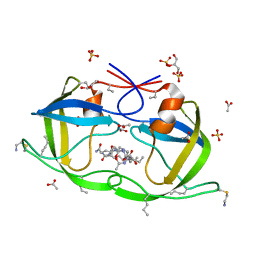 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor MVT-101 | | 分子名称: | ACETATE ION, GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, ... | | 著者 | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | 登録日 | 2006-11-11 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
2JE4
 
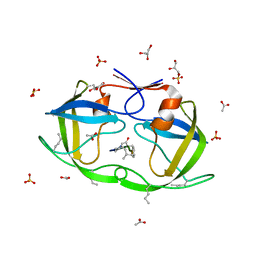 | | Atomic-resolution crystal structure of chemically-synthesized HIV-1 protease in complex with JG-365 | | 分子名称: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | 著者 | Malito, E, Johnson, E.C.B, Tang, W.J. | | 登録日 | 2007-01-15 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease.
J.Am.Chem.Soc., 129, 2007
|
|
2J9J
 
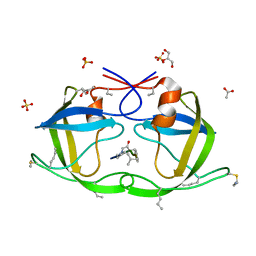 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor JG-365 | | 分子名称: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | 著者 | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | 登録日 | 2006-11-11 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
1C3T
 
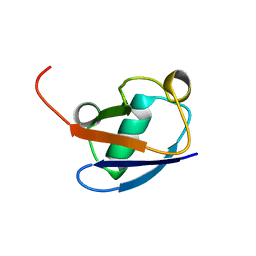 | |
5L2G
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th Position | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5CM)P*GP*CP*G)-3') | | 著者 | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Johnson, E.C, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2016-08-01 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
1B2T
 
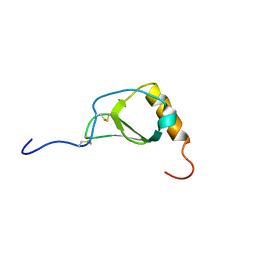 | |
6ALT
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position | | 分子名称: | DNA (5'-D(*(DC5)P*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-08-08 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5L06
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd Position | | 分子名称: | DNA (5'-D(*CP*GP*(5CM)P*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | 著者 | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2016-07-26 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALU
 
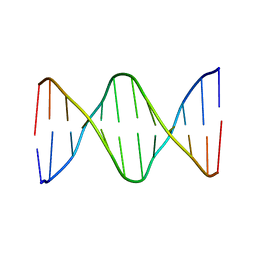 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 8-oxoguanine at the 4th position | | 分子名称: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*CP*GP*CP*(DG3))-3') | | 著者 | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-08-08 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALS
 
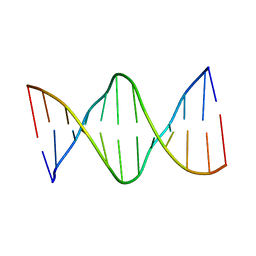 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 4th position | | 分子名称: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | 著者 | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-08-08 | | 公開日 | 2017-09-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5TRN
 
 | | Solution Structure of a DNA Dodecamer with 8-oxoguanine at the 4th position and 5-methylcytosine at the 9th position | | 分子名称: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*G)-3') | | 著者 | Hoppins, J.J, Gruber, D.R, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2016-10-26 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ1
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd position and 8-oxoguanine at the 10th position | | 分子名称: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*CP*(8OG)P*CP*G)-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-02-24 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ3
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th position and 8-oxoguanine at the 10th position | | 分子名称: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-02-24 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ2
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 10th position | | 分子名称: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | 著者 | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | 登録日 | 2017-02-24 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
