3AH8
 
 | | Structure of heterotrimeric G protein Galpha-q beta gamma in complex with an inhibitor YM-254890 | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Nishimura, A, Kitano, K, Takasaki, J, Taniguchi, M, Mizuno, N, Tago, K, Hakoshima, T, Itoh, H. | | 登録日 | 2010-04-20 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
8GZ6
 
 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Nanobody P17 | | 著者 | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | 登録日 | 2022-09-25 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
8GZ5
 
 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P17, ... | | 著者 | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | 登録日 | 2022-09-25 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7C25
 
 | | Glycosidase Wild Type at pH8.0 | | 分子名称: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.505 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C24
 
 | | Glycosidase F290Y at pH8.0 | | 分子名称: | AMMONIUM ION, GLYCEROL, Isomaltose glucohydrolase | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C26
 
 | | Glycosidase Wild Type at pH4.5 | | 分子名称: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
7C27
 
 | | Glycosidase F290Y at pH4.5 | | 分子名称: | AMMONIUM ION, CITRIC ACID, GLYCEROL, ... | | 著者 | Tagami, T, Kikuchi, A, Okuyama, M, Kimura, A. | | 登録日 | 2020-05-07 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural insights reveal the second base catalyst of isomaltose glucohydrolase.
Febs J., 289, 2022
|
|
4JHO
 
 | | Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, GLYCEROL | | 著者 | Tankrathok, A, Luang, S, Robinson, R.C, Kimura, A, Hrmova, M, Ketudat Cairns, J.R. | | 登録日 | 2013-03-05 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural analysis and insights into the glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4JIE
 
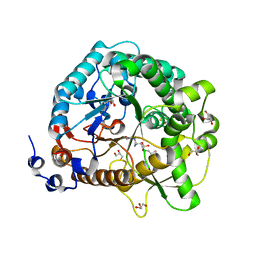 | | Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-mannosidase/beta-glucosidase, GLYCEROL, ... | | 著者 | Tankrathok, A, Luang, S, Robinson, R.C, Kimura, A, Hrmova, M, Ketudat Cairns, J.R. | | 登録日 | 2013-03-05 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural analysis and insights into the glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7EAY
 
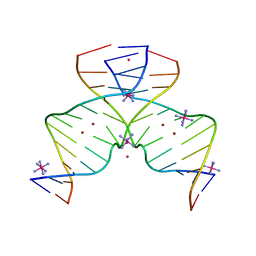 | | DNA containing Cu(II)-mediated 4-N-carboxymethylcytosine base pairs | | 分子名称: | COBALT (II) ION, COBALT HEXAMMINE(III), COPPER (II) ION, ... | | 著者 | Kondo, J, Terashima, A, Yoshimura, A, Tada, Y, Ono, A. | | 登録日 | 2021-03-08 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | DNA containing Cu(II)-mediated 4-N-carboxymethylcytosine base pairs
To Be Published
|
|
8WX5
 
 | |
3WNP
 
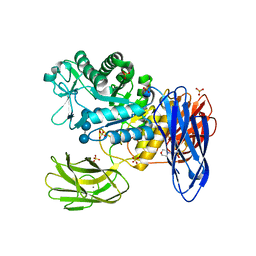 | | D308A, F268V, D469Y, A513V, and Y515S quintuple mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoundecaose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | 著者 | Suzuki, R, Suzuki, N, Fujimoto, Z, Momma, M, Kimura, K, Kitamura, S, Kimura, A, Funane, K. | | 登録日 | 2013-12-10 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular engineering of cycloisomaltooligosaccharide glucanotransferase from Bacillus circulans T-3040: structural determinants for the reaction product size and reactivity.
Biochem.J., 467, 2015
|
|
3WNM
 
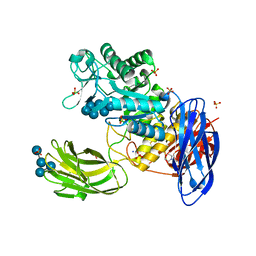 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltoheptaose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | 著者 | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | 登録日 | 2013-12-10 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNL
 
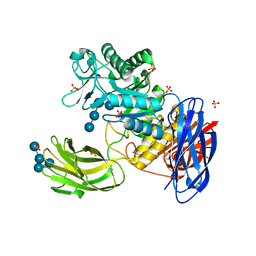 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltohexaose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | 著者 | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | 登録日 | 2013-12-10 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNK
 
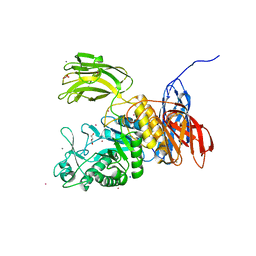 | | Crystal Structure of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase | | 分子名称: | ACETATE ION, CADMIUM ION, CALCIUM ION, ... | | 著者 | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | 登録日 | 2013-12-10 | | 公開日 | 2014-02-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNN
 
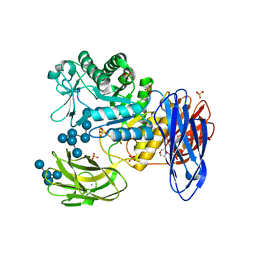 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | 著者 | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | 登録日 | 2013-12-10 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3WNO
 
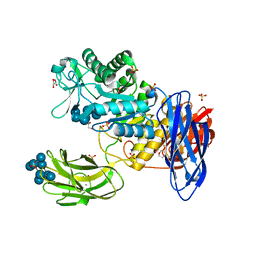 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with cycloisomaltooctaose | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Kobayashi, M, Kimura, A, Funane, K. | | 登録日 | 2013-12-10 | | 公開日 | 2014-02-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
5E1Q
 
 | | Mutant (D415G) GH97 alpha-galactosidase in complex with Gal-Lac | | 分子名称: | CALCIUM ION, GLYCEROL, Retaining alpha-galactosidase, ... | | 著者 | Matsunaga, K, Yamashita, K, Tagami, T, Yao, M, Okuyama, M, Kimura, A. | | 登録日 | 2015-09-30 | | 公開日 | 2016-10-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.943 Å) | | 主引用文献 | Efficient synthesis of alpha-galactosyl oligosaccharides using a mutant Bacteroides thetaiotaomicron retaining alpha-galactosidase (BtGH97b).
FEBS J., 284, 2017
|
|
3KSC
 
 | | Crystal structure of pea prolegumin, an 11S seed globulin from Pisum sativum L. | | 分子名称: | GLYCEROL, LegA class, SULFATE ION | | 著者 | Tandang-Silvas, M.R.G, Fukuda, T, Fukuda, C, Prak, K, Cabanos, C, Kimura, A, Itoh, T, Mikami, B, Maruyama, N, Utsumi, S. | | 登録日 | 2009-11-21 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.606 Å) | | 主引用文献 | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
4Y7E
 
 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus with mannohexaose | | 分子名称: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | 著者 | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | 登録日 | 2015-02-14 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
5X7S
 
 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase, terbium derivative | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | 登録日 | 2017-02-27 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7Q
 
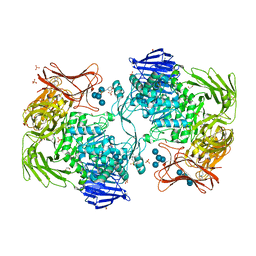 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase complexed with maltohexaose | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | 著者 | Fujimoto, Z, Kishine, N, Suzuki, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | 登録日 | 2017-02-27 | | 公開日 | 2017-07-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
5X7O
 
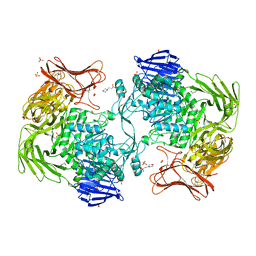 | | Crystal structure of Paenibacillus sp. 598K alpha-1,6-glucosyltransferase | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Fujimoto, Z, Suzuki, N, Kishine, N, Momma, M, Ichinose, H, Kimura, A, Funane, K. | | 登録日 | 2017-02-27 | | 公開日 | 2017-07-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Carbohydrate-binding architecture of the multi-modular alpha-1,6-glucosyltransferase from Paenibacillus sp. 598K, which produces alpha-1,6-glucosyl-alpha-glucosaccharides from starch
Biochem. J., 474, 2017
|
|
