4AU8
 
 | | Crystal structure of compound 4a in complex with cdk5, showing an unusual binding mode to the hinge region via a water molecule | | 分子名称: | 4-(1,3-benzothiazol-2-yl)thiophene-2-sulfonamide, CYCLIN-DEPENDENT KINASE 5, IMIDAZOLE, ... | | 著者 | Malmstrom, J, Viklund, J, Slivo, C, Costa, A, Maudet, M, Sandelin, C, Hiller, G, Olsson, L.L, Aagaard, A, Geschwindner, S, Xue, Y, Vasange, M. | | 登録日 | 2012-05-14 | | 公開日 | 2013-03-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Synthesis and Structure-Activity Relationship of 4-(1,3-Benzothiazol-2-Yl)-Thiophene-2-Sulfonamides as Cyclin-Dependent Kinase 5 (Cdk5)/P25 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8R1X
 
 | |
6I1S
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with FKBP12 and the inhibitor E6201 | | 分子名称: | (4~{S},5~{R},6~{Z},9~{S},10~{S},12~{E})-16-(ethylamino)-4,5-dimethyl-9,10,18-tris(oxidanyl)-3-oxabicyclo[12.4.0]octadeca-1(14),6,12,15,17-pentaene-2,8-dione, 1,2-ETHANEDIOL, Activin receptor type-1, ... | | 著者 | Williams, E.P, Pinkas, D.M, Fortin, J, Newman, J.A, Bradshaw, W.J, Mahajan, P, Kupinska, K, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | 登録日 | 2018-10-30 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Mutant ACVR1 Arrests Glial Cell Differentiation to Drive Tumorigenesis in Pediatric Gliomas.
Cancer Cell, 37, 2020
|
|
4LI5
 
 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | 分子名称: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | 著者 | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | 登録日 | 2013-07-02 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
8EUA
 
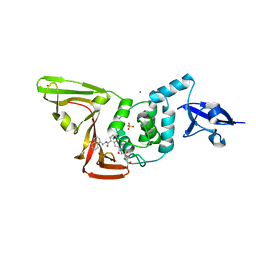 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | 分子名称: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | 著者 | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | 登録日 | 2022-10-18 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | 分子名称: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-07 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7SI9
 
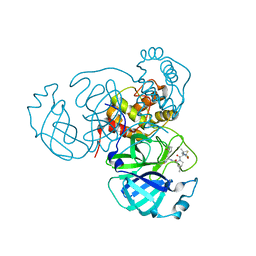 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-10-12 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
2ORR
 
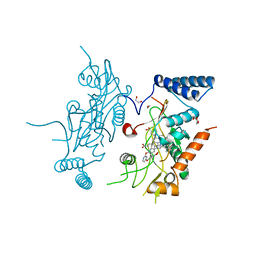 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (Delta 114) 4-(Benzo[1,3]dioxol-5-yloxy)-2-(4-imidazol-1-yl-phenoxy)-pyrimidine Complex | | 分子名称: | 1,2-ETHANEDIOL, 4-(1,3-BENZODIOXOL-5-YLOXY)-2-[4-(1H-IMIDAZOL-1-YL)PHENOXY]PYRIMIDINE, Nitric oxide synthase, ... | | 著者 | Adler, M, Whitlow, M. | | 登録日 | 2007-02-04 | | 公開日 | 2007-04-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
2ORT
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (Delta 114) 1-Benzo[1,3]dioxol-5-ylmethyl-3S-(4-imidazol-1-yl-phenoxy)-piperidine Complex | | 分子名称: | (3S)-1-(1,3-BENZODIOXOL-5-YLMETHYL)-3-[4-(1H-IMIDAZOL-1-YL)PHENOXY]PIPERIDINE, Nitric oxide synthase, inducible, ... | | 著者 | Adler, M, Whitlow, M. | | 登録日 | 2007-02-04 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
2ORQ
 
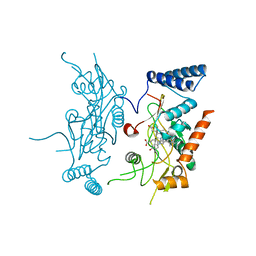 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (DELTA 114) 4-(imidazol-1-yl)phenol and piperonylamine Complex | | 分子名称: | 1-(1,3-BENZODIOXOL-5-YL)METHANAMINE, 4-(1H-IMIDAZOL-1-YL)PHENOL, Nitric oxide synthase, ... | | 著者 | Adler, M, Whitlow, M. | | 登録日 | 2007-02-04 | | 公開日 | 2007-04-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
2ORS
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (DELTA 114) 4-(Benzo[1,3]dioxol-5-yloxy)-2-(4-imidazol-1-yl-phenoxy)-6-methyl-pyrimidine Complex | | 分子名称: | 1,2-ETHANEDIOL, 4-(1,3-BENZODIOXOL-5-YLOXY)-2-[4-(1H-IMIDAZOL-1-YL)PHENOXY]-6-METHYLPYRIMIDINE, Nitric oxide synthase, ... | | 著者 | Adler, M, Whitlow, M. | | 登録日 | 2007-02-04 | | 公開日 | 2007-04-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
6N9T
 
 | | Structure of a peptide-based photo-affinity cross-linker with Herceptin Fc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin G1 FC, ... | | 著者 | Sadowsky, J, Ultsch, M, Vance, N, Wang, W. | | 登録日 | 2018-12-04 | | 公開日 | 2019-01-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.576 Å) | | 主引用文献 | Development, Optimization, and Structural Characterization of an Efficient Peptide-Based Photoaffinity Cross-Linking Reaction for Generation of Homogeneous Conjugates from Wild-Type Antibodies.
Bioconjug. Chem., 30, 2019
|
|
7TWF
 
 | |
7TWQ
 
 | |
7TWI
 
 | |
7TX5
 
 | |
7TWG
 
 | |
7TWS
 
 | |
7TWR
 
 | |
7TWP
 
 | |
7TWH
 
 | |
7TWJ
 
 | |
7TX3
 
 | |
7TWN
 
 | |
7TWO
 
 | |
