4V1O
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | 分子名称: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | 著者 | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | 登録日 | 2014-09-29 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9.7 Å) | | 主引用文献 | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4V1M
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | 分子名称: | 5'-D(*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP)-3', 5'-D(*CP*CP*AP*GP*GP*AP)-3', 5'-D(*TP*CP*AP*AP*GP*TP*AP*CP*TP*TP*TP*TP*TP*CP *CP*BRUP*GP*GP*TP*C)-3', ... | | 著者 | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | 登録日 | 2014-09-29 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
4V1N
 
 | | Architecture of the RNA polymerase II-Mediator core transcription initiation complex | | 分子名称: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | 著者 | Plaschka, C, Lariviere, L, Wenzeck, L, Hemann, M, Tegunov, D, Petrotchenko, E.V, Borchers, C.H, Baumeister, W, Herzog, F, Villa, E, Cramer, P. | | 登録日 | 2014-09-29 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Architecture of the RNA Polymerase II-Mediator Core Initiation Complex.
Nature, 518, 2015
|
|
5FLM
 
 | | Structure of transcribing mammalian RNA polymerase II | | 分子名称: | DNA, DNA-RNA ELONGATION SCAFFOLD, DNA-DIRECTED RNA POLYMERASE, ... | | 著者 | Bernecky, C, Herzog, F, Baumeister, W, Plitzko, J.M, Cramer, P. | | 登録日 | 2015-10-26 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of Transcribing Mammalian RNA Polymerase II
Nature, 529, 2016
|
|
3TJ1
 
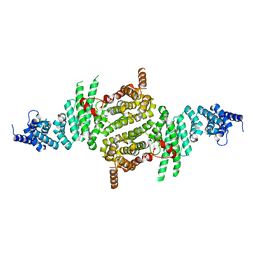 | | Crystal Structure of RNA Polymerase I Transcription Initiation Factor Rrn3 | | 分子名称: | RNA polymerase I-specific transcription initiation factor RRN3 | | 著者 | Blattner, C, Jennebach, S, Herzog, F, Mayer, A, Cheung, A.C.M, Witte, G, Lorenzen, K, Hopfner, K.-P, Heck, A.J.R, Aebersold, R, Cramer, P. | | 登録日 | 2011-08-23 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth.
Genes Dev., 25, 2011
|
|
7ZKE
 
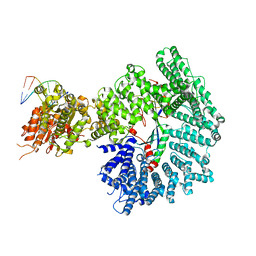 | | Mot1:TBP:DNA - pre-hydrolysis state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (36-MER), ... | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-04-12 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z8S
 
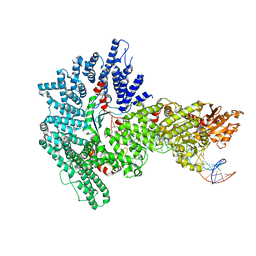 | | Mot1:TBP:DNA - post hydrolysis state | | 分子名称: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-03-18 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z7N
 
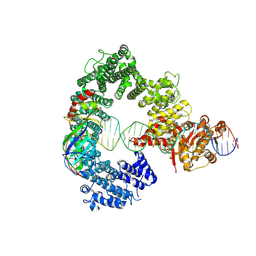 | | Mot1E1434Q:TBP:DNA - substrate recognition state | | 分子名称: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-03-16 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZB5
 
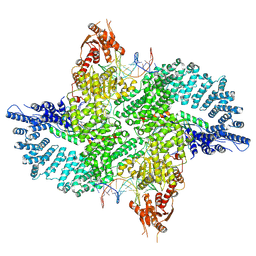 | | Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer | | 分子名称: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | 著者 | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | 登録日 | 2022-03-23 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5VQ9
 
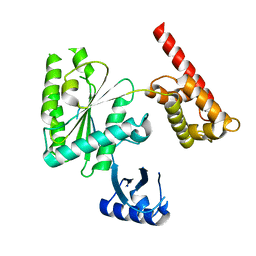 | | Structure of human TRIP13, Apo form | | 分子名称: | Pachytene checkpoint protein 2 homolog | | 著者 | Ye, Q, Corbett, K.D. | | 登録日 | 2017-05-08 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
5VQA
 
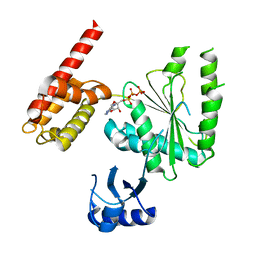 | | Structure of human TRIP13, ATP-bound form | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | 著者 | Ye, Q, Corbett, K.D. | | 登録日 | 2017-05-08 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
6DD9
 
 | | Structure of mouse SYCP3, P1 form | | 分子名称: | Synaptonemal complex protein 3 | | 著者 | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | 登録日 | 2018-05-09 | | 公開日 | 2018-08-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
6ZCE
 
 | | Structure of a yeast ABCE1-bound 43S pre-initiation complex | | 分子名称: | 18S ribosomal RNA (1719-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-06-10 | | 公開日 | 2020-10-07 | | 最終更新日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (5.3 Å) | | 主引用文献 | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
6ZU9
 
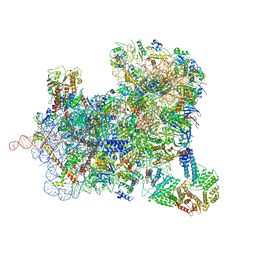 | | Structure of a yeast ABCE1-bound 48S initiation complex | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-22 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
5D4W
 
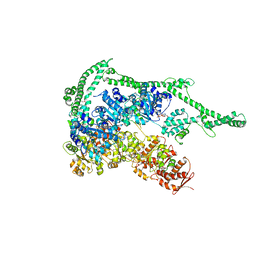 | |
4OUC
 
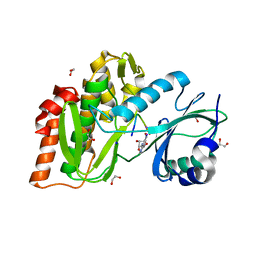 | | Structure of human haspin in complex with histone H3 substrate | | 分子名称: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, Histone H3.2, ... | | 著者 | Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2014-02-15 | | 公開日 | 2014-04-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Modulation of the chromatin phosphoproteome by the haspin protein kinase.
Mol Cell Proteomics, 13, 2014
|
|
4P0T
 
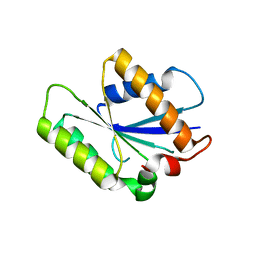 | |
6N88
 
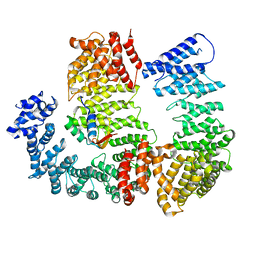 | |
6N89
 
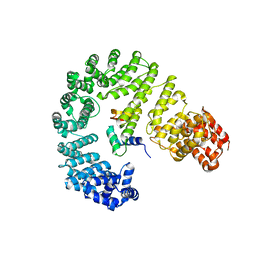 | |
6DD8
 
 | | Structure of mouse SYCP3, P21 form | | 分子名称: | Synaptonemal complex protein 3 | | 著者 | Rosenberg, S.C, Munoz, I.C, Uson, I, Corbett, K.D. | | 登録日 | 2018-05-09 | | 公開日 | 2018-08-01 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A conserved filamentous assembly underlies the structure of the meiotic chromosome axis.
Elife, 8, 2019
|
|
5L3X
 
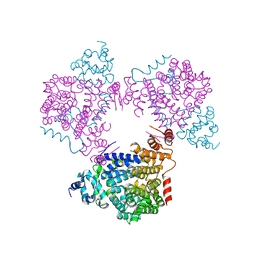 | |
5LSJ
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C delta-HEAD2 COMPLEX | | 分子名称: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | 著者 | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | 登録日 | 2016-09-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | 分子名称: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | 著者 | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | 登録日 | 2016-09-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.502 Å) | | 主引用文献 | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | 分子名称: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | 著者 | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | 登録日 | 2016-09-02 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
6ZVJ
 
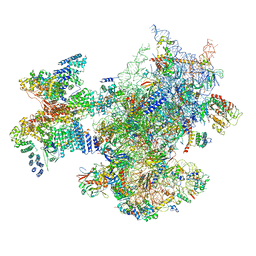 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State II | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-24 | | 公開日 | 2020-10-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
