2ND5
 
 | | Lysine dimethylated FKBP12 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase FKBP1A | | 著者 | Hattori, Y, Sebera, J, Sychrovsky, V, Furuita, K, Sugiki, T, Ohki, I, Ikegami, T, Kobayashi, N, Tanaka, Y, Fujiwara, T, Kojima, C. | | 登録日 | 2016-05-05 | | 公開日 | 2017-05-17 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Observation of Protein Surface Salt Bridges at Neutral pH
To be Published
|
|
5IX7
 
 | | Crystal structure of metallo-DNA nanowire with infinite one-dimensional silver array | | 分子名称: | DNA (5'-D(*GP*GP*AP*CP*TP*(CBR)P*GP*AP*CP*TP*CP*C)-3'), POTASSIUM ION, SILVER ION | | 著者 | Kondo, J, Tada, Y, Dairaku, T, Hattori, Y, Saneyoshi, H, Ono, A, Tanaka, Y. | | 登録日 | 2016-03-23 | | 公開日 | 2017-07-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.398 Å) | | 主引用文献 | A metallo-DNA nanowire with uninterrupted one-dimensional silver array
Nat Chem, 9, 2017
|
|
3AW0
 
 | | Structure of SARS 3CL protease with peptidic aldehyde inhibitor | | 分子名称: | 3C-Like Proteinase, peptide ACE-SER-ALA-VAL-LEU-HIS-H | | 著者 | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | 登録日 | 2011-03-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3AW1
 
 | | Structure of SARS 3CL protease auto-proteolysis resistant mutant in the absent of inhibitor | | 分子名称: | 3C-Like Proteinase | | 著者 | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | 登録日 | 2011-03-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3ATW
 
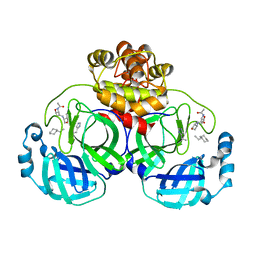 | | Structure-Based Design, Synthesis, Evaluation of Peptide-mimetic SARS 3CL Protease Inhibitors | | 分子名称: | 3C-Like Proteinase, peptide ACE-THR-VAL-ALC-HIS-H | | 著者 | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | 登録日 | 2011-01-20 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3AVZ
 
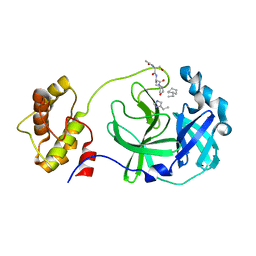 | | Structure of SARS 3CL protease with peptidic aldehyde inhibitor containing cyclohexyl side chain | | 分子名称: | 3C-Like Proteinase, peptide ACE-SER-ALA-VAL-ALC-HIS-H | | 著者 | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | 登録日 | 2011-03-09 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
7XW8
 
 | |
4TRW
 
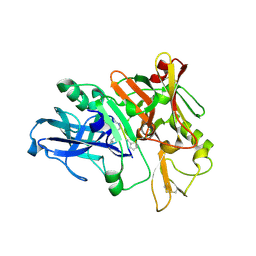 | | Structure of BACE1 complex with a syn-HEA-type inhibitor | | 分子名称: | Beta-secretase 1, L-alpha-glutamyl-L-isoleucyl-N-[(2R,3S)-1-{[(1S)-1-carboxybutyl]amino}-2-hydroxy-5-methylhexan-3-yl]-3-thiophen-2-yl-L-alaninamide | | 著者 | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-06-18 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TRZ
 
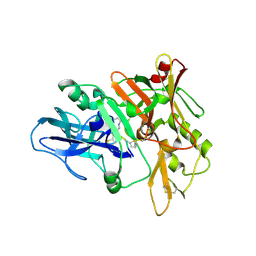 | | Structure of BACE1 complex with 2-thiophenyl HEA-type inhibitor | | 分子名称: | 2-thiophenyl HEA-type inhibitor, Beta-secretase 1 | | 著者 | Akaji, K, Teruya, K, Akiyama, T, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-06-18 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Evaluation of transition-state mimics in a superior BACE1 cleavage sequence as peptide-mimetic BACE1 inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TWW
 
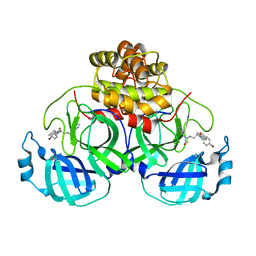 | | Structure of SARS-3CL protease complex with a Bromobenzoyl (S,R)-N-decalin type inhibitor | | 分子名称: | (2S)-2-({[(3S,4aR,8aS)-2-(4-bromobenzoyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | 著者 | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-07-02 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TWY
 
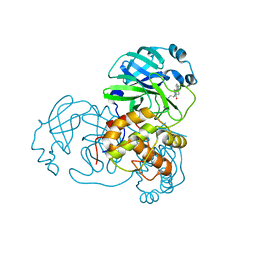 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | 分子名称: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | 著者 | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-07-02 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
3GP7
 
 | |
3NBT
 
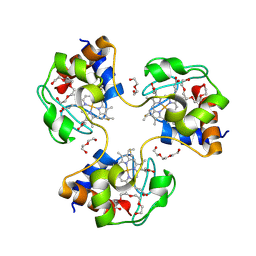 | | Crystal structure of trimeric cytochrome c from horse heart | | 分子名称: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | 著者 | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | 登録日 | 2010-06-04 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NBS
 
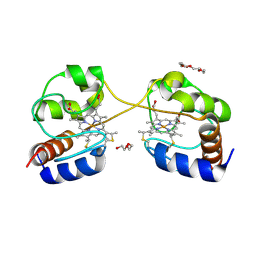 | | Crystal structure of dimeric cytochrome c from horse heart | | 分子名称: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | 著者 | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | 登録日 | 2010-06-04 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2RUJ
 
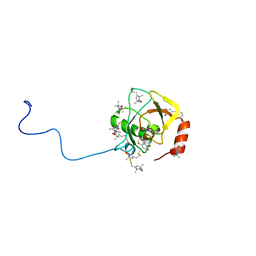 | | Solution structure of MTSL spin-labeled Schizosaccharomyces pombe Sin1 CRIM domain | | 分子名称: | Stress-activated map kinase-interacting protein 1 | | 著者 | Furuita, K, Kataoka, S, Sugiki, T, Kobayashi, N, Ikegami, T, Shiozaki, K, Fujiwara, T, Kojima, C. | | 登録日 | 2014-07-24 | | 公開日 | 2015-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Utilization of paramagnetic relaxation enhancements for high-resolution NMR structure determination of a soluble loop-rich protein with sparse NOE distance restraints
J.Biomol.Nmr, 61, 2015
|
|
4WY3
 
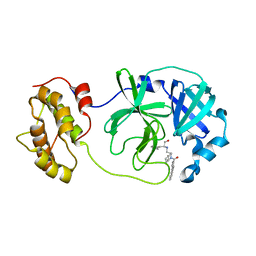 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor | | 分子名称: | (2S)-2-({[(3R,4aS,8aR)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | 著者 | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | 登録日 | 2014-11-15 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
7E0G
 
 | |
