6DCM
 
 | |
8E91
 
 | |
8E8Q
 
 | | Cryo-EM structure of substrate-free DNClpX.ClpP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | 著者 | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | 登録日 | 2022-08-25 | | 公開日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (3.12 Å) | | 主引用文献 | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8E7V
 
 | | Cryo-EM structure of substrate-free DNClpX.ClpP from singly capped particles | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpX, ... | | 著者 | Ghanbarpour, A, Cohen, S, Davis, J.H, Sauer, R.T. | | 登録日 | 2022-08-24 | | 公開日 | 2023-11-08 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structure of substrate-free DNClpX.ClpP
Nat Commun, 2023
|
|
8KB4
 
 | | Cryo-EM structure of human TMEM87A A308M | | 分子名称: | (9R,12R)-15-amino-12-hydroxy-6,12-dioxo-7,11,13-trioxa-12lambda~5~-phosphapentadecan-9-yl undecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 87A,EGFP | | 著者 | Han, A, Kim, H.M. | | 登録日 | 2023-08-03 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structure of human TMEM87A, PE-bound
To Be Published
|
|
8HTT
 
 | | Cryo-EM structure of human TMEM87A, gluconate-bound | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-gluconic acid, Transmembrane protein 87A,EGFP, ... | | 著者 | Han, A, Kim, H.M. | | 登録日 | 2022-12-21 | | 公開日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of human TMEM87A, PE-bound
To Be Published
|
|
8HSI
 
 | | Cryo-EM structure of human TMEM87A, PE-bound | | 分子名称: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Han, A, Kim, H.M. | | 登録日 | 2022-12-19 | | 公開日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Cryo-EM structure of human TMEM87A, PE-bound
To Be Published
|
|
1N6J
 
 | | Structural basis of sequence-specific recruitment of histone deacetylases by Myocyte Enhancer Factor-2 | | 分子名称: | 5'-D(*AP*GP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*CP*T)-3', Calcineurin-binding protein Cabin 1, ... | | 著者 | Han, A, Pan, F, Stroud, J.C, Youn, H.D, Liu, J.O, Chen, L. | | 登録日 | 2002-11-11 | | 公開日 | 2003-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2
Nature, 422, 2003
|
|
6WP0
 
 | |
6ON5
 
 | |
6ON8
 
 | |
6ON7
 
 | |
1HRI
 
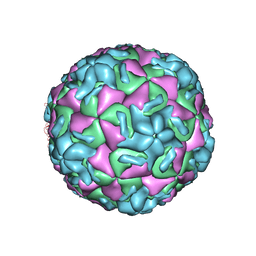 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | 分子名称: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | 著者 | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | 登録日 | 1992-10-01 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
8GR3
 
 | |
8GR1
 
 | |
6WP2
 
 | |
5ALB
 
 | | Ticagrelor antidote candidate MEDI2452 in complex with ticagrelor | | 分子名称: | MEDI2452 HEAVY CHAIN, MEDI2452 LIGHT CHAIN, Ticagrelor | | 著者 | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | 登録日 | 2015-03-07 | | 公開日 | 2015-04-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
7N40
 
 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | 分子名称: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | 著者 | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | 登録日 | 2021-06-02 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
6WNF
 
 | |
6WP1
 
 | |
6WNJ
 
 | |
3PIQ
 
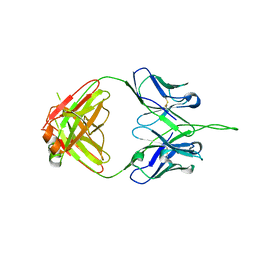 | | Crystal structure of human 2909 Fab, a quaternary structure-specific antibody against HIV-1 | | 分子名称: | Human monoclonal antibody 2909 Fab heavy chain, Human monoclonal antibody 2909 Fab light chain | | 著者 | Changela, A, Gorny, M.K, Zolla-Pazner, S, Kwong, P.D. | | 登録日 | 2010-11-07 | | 公開日 | 2011-01-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.325 Å) | | 主引用文献 | Crystal Structure of Human Antibody 2909 Reveals Conserved Features of Quaternary Structure-Specific Antibodies That Potently Neutralize HIV-1.
J.Virol., 85, 2011
|
|
5ALC
 
 | | Ticagrelor antidote candidate Fab 72 in complex with ticagrelor | | 分子名称: | ANTI-TICAGRELOR FAB 72, HEAVY CHAIN, LIGHT CHAIN, ... | | 著者 | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | 登録日 | 2015-03-07 | | 公開日 | 2015-04-01 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
3IAA
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP bound form | | 分子名称: | CalG2, THYMIDINE-5'-DIPHOSPHATE | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2009-07-13 | | 公開日 | 2010-06-02 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.505 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3IA7
 
 | | Crystal Structure of CalG4, the Calicheamicin Glycosyltransferase | | 分子名称: | CALCIUM ION, CHLORIDE ION, CalG4 | | 著者 | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | 登録日 | 2009-07-13 | | 公開日 | 2010-06-02 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
