8IIC
 
 | |
8IIB
 
 | | Crystal structure of Israeli acute paralysis virus RNA-dependent RNA polymerase delta85 mutant (residues 86-546) | | 分子名称: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Fang, X, Lu, G, Hou, C, Gong, P. | | 登録日 | 2023-02-24 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Unusual substructure conformations observed in crystal structures of a dicistrovirus RNA-dependent RNA polymerase suggest contribution of the N-terminal extension in proper folding.
Virol Sin, 38, 2023
|
|
6NXJ
 
 | | Flavin Transferase ApbE from Vibrio cholerae, H257G mutant | | 分子名称: | FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | 著者 | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-02-08 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6NXI
 
 | | Flavin Transferase ApbE from Vibrio cholerae | | 分子名称: | 1,2-ETHANEDIOL, FAD:protein FMN transferase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Osipiuk, J, Fang, X, Chakravarthy, S, Juarez, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-02-08 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Conserved residue His-257 ofVibrio choleraeflavin transferase ApbE plays a critical role in substrate binding and catalysis.
J.Biol.Chem., 294, 2019
|
|
6JIQ
 
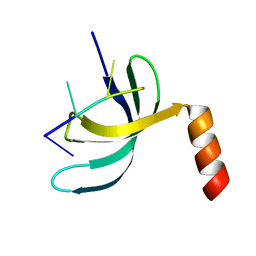 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), SP_0782 | | 著者 | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | 登録日 | 2019-02-22 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6JIP
 
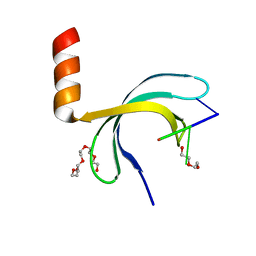 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT6 | | 分子名称: | DNA (5'-D(*TP*TP*TP*TP*T)-3'), PENTAETHYLENE GLYCOL, SP_0782 | | 著者 | Fang, X, Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | 登録日 | 2019-02-22 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.659 Å) | | 主引用文献 | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6M3A
 
 | |
8JY0
 
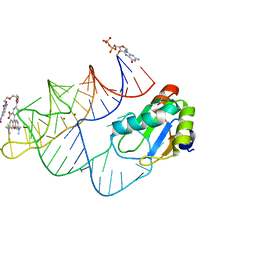 | | Crystal structure of RhoBAST complexed with TMR-DN | | 分子名称: | 2,4-dinitroaniline, 5-aminocarbonyl-2-[3-(dimethylamino)-6-dimethylazaniumylidene-xanthen-9-yl]benzoate, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Zhang, Y, Xiao, Y, Xu, Z, Fang, X. | | 登録日 | 2023-07-02 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural mechanisms for binding and activation of a contact-quenched fluorophore by RhoBAST.
Nat Commun, 15, 2024
|
|
2LF0
 
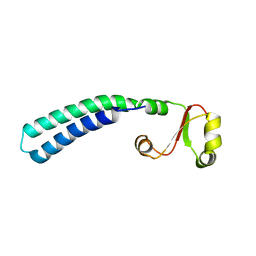 | | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering, NESG target SfR339/OCSP target sf3636 | | 分子名称: | Uncharacterized protein yibL | | 著者 | Wu, B, Lemak, A, Yee, A, Lee, H, Gutmanas, A, Semesi, A, Garcia, M, Fang, X, Wang, Y, Prestegard, J.H, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | 登録日 | 2011-06-27 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Solution structure of sf3636, a two-domain unknown function protein from Shigella flexneri 2a, determined by joint refinement of NMR, residual dipolar couplings and small-angle X-ray scattering
To be Published
|
|
7WIA
 
 | |
7WII
 
 | | The THF-II riboswitch bound to NPR | | 分子名称: | 2-AMINO-7,8-DIHYDRO-6-(1,2,3-TRIHYDROXYPROPYL)-4(1H)-PTERIDINONE, RNA (50-MER) | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WI9
 
 | | The THF-II riboswitch bound to THF and soaking with SeUrea | | 分子名称: | (6S)-5,6,7,8-TETRAHYDROFOLATE, RNA (50-MER), selenourea | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WIB
 
 | | The THF-II riboswitch bound to THF | | 分子名称: | (6S)-5,6,7,8-TETRAHYDROFOLATE, RNA (50-MER) | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WIF
 
 | | The THF-II riboswitch bound to H4B | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, RNA (50-MER) | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.86 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
7WIE
 
 | | The THF-II riboswitch bound to 7DG | | 分子名称: | 7-DEAZAGUANINE, RNA (50-MER) | | 著者 | Xu, L, Fang, X, Xiao, Y. | | 登録日 | 2022-01-03 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural insights into translation regulation by the THF-II riboswitch.
Nucleic Acids Res., 51, 2023
|
|
1E5C
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | 分子名称: | XYLANASE D | | 著者 | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | 登録日 | 2000-07-24 | | 公開日 | 2001-05-25 | | 最終更新日 | 2018-10-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
1E5B
 
 | | Internal xylan binding domain from C. fimi Xyn10A, R262G mutant | | 分子名称: | XYLANASE D | | 著者 | Simpson, P.J, Hefang, X, Bolam, D.N, Gilbert, H.J, Williamson, M.P. | | 登録日 | 2000-07-24 | | 公開日 | 2001-05-25 | | 最終更新日 | 2018-10-24 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Structural Basis for the Ligand Specificity of Family 2 Carbohydrate Binding Nodules
J.Biol.Chem., 275, 2000
|
|
1Y9X
 
 | |
4M1P
 
 | |
7C7U
 
 | | Biofilm associated protein - BSP domain | | 分子名称: | Biofilm-associated surface protein, CALCIUM ION | | 著者 | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | 登録日 | 2020-05-26 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
7C7R
 
 | | Biofilm associated protein - B domain | | 分子名称: | Biofilm-associated surface protein, CALCIUM ION | | 著者 | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | 登録日 | 2020-05-26 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
5T16
 
 | |
8KFA
 
 | | Cryo-EM structure of HSV-1 gB with D48 Fab complex | | 分子名称: | D48 heavy chain, D48 light chain, Envelope glycoprotein B | | 著者 | Yang, J, Sun, C, Fang, X, Zeng, M, Liu, Z. | | 登録日 | 2023-08-15 | | 公開日 | 2024-01-03 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | The structure of HSV-1 gB bound to a potent neutralizing antibody reveals a conservative antigenic domain across herpesviruses
hlife, 2023
|
|
3SPF
 
 | | Crystal Structure of Bcl-xL bound to BM501 | | 分子名称: | 4-(4-chlorophenyl)-1-[(3S)-3,4-dihydroxybutyl]-N-[3-(4-methylpiperazin-1-yl)propyl]-3-phenyl-1H-pyrrole-2-carboxamide, Bcl-2-like protein 1, GLYCEROL | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2011-07-01 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Design of Bcl-2 and Bcl-xL Inhibitors with Subnanomolar Binding Affinities Based upon a New Scaffold.
J.Med.Chem., 55, 2012
|
|
8FP3
 
 | | PKCeta kinase domain in complex with compound 11 | | 分子名称: | (3P)-3-{4-[(3R,5S)-3-amino-5-methylpiperidin-1-yl]-6-chloro-7H-pyrrolo[2,3-d]pyrimidin-5-yl}benzonitrile, Protein kinase C eta type | | 著者 | Johnson, E. | | 登録日 | 2023-01-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design and Synthesis of Functionally Active 5-Amino-6-Aryl Pyrrolopyrimidine Inhibitors of Hematopoietic Progenitor Kinase 1.
J.Med.Chem., 66, 2023
|
|
