2FFL
 
 | |
2QVW
 
 | |
6MCC
 
 | |
5I4A
 
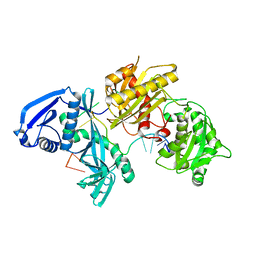 | | X-ray crystal structure of Marinitoga piezophila Argonaute in complex with 5' OH guide RNA | | 分子名称: | Argonaute protein, RNA (5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*U)-3') | | 著者 | Doxzen, K.W, Kaya, E, Knoll, K.R, Wilson, R.C, Strutt, S.C, Kranzusch, P.J, Doudna, J.A. | | 登録日 | 2016-02-11 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | A bacterial Argonaute with noncanonical guide RNA specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4ZT9
 
 | |
4ZT0
 
 | |
7S37
 
 | | Cas9:sgRNA (S. pyogenes) in the open-protein conformation | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Single-guide RNA | | 著者 | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | 登録日 | 2021-09-04 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S38
 
 | | Cas9:sgRNA:DNA (S. pyogenes) forming a 3-base-pair R-loop | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | 著者 | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | 登録日 | 2021-09-04 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S3H
 
 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, open-protein/linear-DNA conformation | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | 著者 | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | 登録日 | 2021-09-06 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S36
 
 | | Cas9:sgRNA:DNA (S. pyogenes) with 0 RNA:DNA base pairs, closed-protein/bent-DNA conformation | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, Non-target DNA strand, Single-guide RNA, ... | | 著者 | Cofsky, J.C, Soczek, K.M, Knott, G.J, Nogales, E, Doudna, J.A. | | 登録日 | 2021-09-04 | | 公開日 | 2022-04-20 | | 最終更新日 | 2022-05-04 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | CRISPR-Cas9 bends and twists DNA to read its sequence.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5W1H
 
 | |
5VVK
 
 | | Cas1-Cas2 bound to full-site mimic | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*CP*CP*AP*CP*CP*AP*GP*TP*G)-3'), ... | | 著者 | Wright, A.V, Knott, G.J, Doxzen, K.D, Doudna, J.A. | | 登録日 | 2017-05-19 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5VVJ
 
 | | Cas1-Cas2 bound to half-site intermediate | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (112-MER), ... | | 著者 | Wright, A.V, Knott, G.J, Doxzen, K.W, Doudna, J.A. | | 登録日 | 2017-05-19 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.89 Å) | | 主引用文献 | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5W1I
 
 | |
5WFE
 
 | | Cas1-Cas2-IHF-DNA holo-complex | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | 著者 | Wright, A.V, Liu, J.J, Nogales, E, Doudna, J.A. | | 登録日 | 2017-07-11 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.64 Å) | | 主引用文献 | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5WLH
 
 | |
8VXY
 
 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | 著者 | Tuck, O.T, Hu, J.J, Doudna, J.A. | | 登録日 | 2024-02-06 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.19 Å) | | 主引用文献 | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8VX9
 
 | |
8VXA
 
 | |
8VXC
 
 | |
8UZB
 
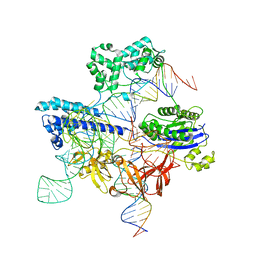 | | Cryo-EM structure of iGeoCas9 in complex with sgRNA and target DNA | | 分子名称: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, RNA (107-MER), ... | | 著者 | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
8UZA
 
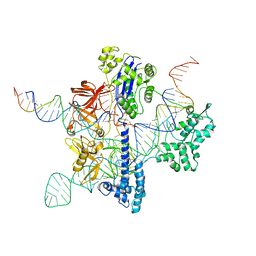 | | Cryo-EM structure of GeoCas9 in complex with sgRNA and target DNA | | 分子名称: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, Target strand DNA, ... | | 著者 | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
4WYQ
 
 | |
7THB
 
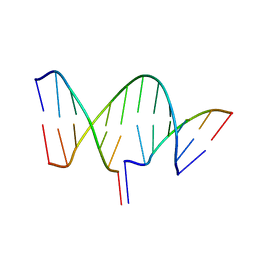 | | Crystal structure of an RNA-5'/DNA-3' strand exchange junction | | 分子名称: | DNA (5'-D(*GP*AP*TP*GP*CP*TP*C)-3'), DNA (5'-D(*GP*TP*AP*AP*GP*CP*AP*GP*CP*AP*TP*C)-3'), RNA (5'-R(*AP*GP*CP*UP*UP*AP*C)-3') | | 著者 | Cofsky, J.C, Knott, G.J, Gee, C.L, Doudna, J.A. | | 登録日 | 2022-01-10 | | 公開日 | 2022-04-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Crystal structure of an RNA/DNA strand exchange junction.
Plos One, 17, 2022
|
|
1GID
 
 | | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | | 分子名称: | COBALT HEXAMMINE(III), MAGNESIUM ION, P4-P6 RNA RIBOZYME DOMAIN | | 著者 | Cate, J.H, Gooding, A.R, Podell, E, Zhou, K, Golden, B.L, Kundrot, C.E, Cech, T.R, Doudna, J.A. | | 登録日 | 1996-08-22 | | 公開日 | 1996-12-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of a group I ribozyme domain: principles of RNA packing.
Science, 273, 1996
|
|
