2A1E
 
 | | High resolution structure of HIV-1 PR with TS-126 | | 分子名称: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Demitri, N, Geremia, S, Randaccio, L, Wuerges, J, Benedetti, F, Berti, F, Dinon, F, Campaner, P, Tell, G. | | 登録日 | 2005-06-20 | | 公開日 | 2006-02-21 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | A potent HIV protease inhibitor identified in an epimeric mixture by high-resolution protein crystallography.
Chemmedchem, 1, 2006
|
|
6Z4N
 
 | | CRYSTAL STRUCTURE OF OASS COMPLEXED WITH UPAR INHIBITOR | | 分子名称: | (1~{S},2~{S})-1-[(4-methylphenyl)methyl]-2-phenyl-cyclopropane-1-carboxylic acid, COBALT (II) ION, Cysteine synthase A, ... | | 著者 | Demitri, N, Storici, P, Campanini, B. | | 登録日 | 2020-05-25 | | 公開日 | 2021-04-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Investigational Studies on a Hit Compound Cyclopropane-Carboxylic Acid Derivative Targeting O -Acetylserine Sulfhydrylase as a Colistin Adjuvant.
Acs Infect Dis., 7, 2021
|
|
6EGR
 
 | | Crystal structure of Citrobacter freundii methionine gamma-lyase with V358Y replacement | | 分子名称: | DI(HYDROXYETHYL)ETHER, Methionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Revtovich, S.V, Demitri, N, Raboni, S, Nikulin, A.D, Morozova, E.A, Demidkina, T.V, Storici, P, Mozzarelli, A. | | 登録日 | 2017-09-12 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Engineering methionine gamma-lyase from Citrobacter freundii for anticancer activity.
Biochim Biophys Acta Proteins Proteom, 1866, 2018
|
|
3K4V
 
 | | New crystal form of HIV-1 Protease/Saquinavir structure reveals carbamylation of N-terminal proline | | 分子名称: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Olajuyigbe, F.M, Demitri, N, Ajele, J.O, Maurizio, E, Randaccio, L, Geremia, S. | | 登録日 | 2009-10-06 | | 公開日 | 2010-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Carbamylation of N-terminal proline.
ACS Med Chem Lett, 1, 2010
|
|
6RV5
 
 | | X-ray structure of the levansucrase from Erwinia tasmaniensis in complex with levanbiose | | 分子名称: | GLYCEROL, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION, ... | | 著者 | Polsinelli, I, Caliandro, R, Demitri, N, Benini, S. | | 登録日 | 2019-05-31 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | The Structure of Sucrose-Soaked Levansucrase Crystals fromErwinia tasmaniensisreveals a Binding Pocket for Levanbiose.
Int J Mol Sci, 21, 2019
|
|
5FR7
 
 | | Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | 分子名称: | AMYR | | 著者 | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | 登録日 | 2015-12-16 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
5FRK
 
 | | SeMet crystal structure of Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | 分子名称: | AMYR | | 著者 | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | 登録日 | 2015-12-18 | | 公開日 | 2017-02-15 | | 最終更新日 | 2019-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
7QF6
 
 | |
7QQE
 
 | | Nuclear factor one X - NFIX in P41212 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nuclear factor 1 X-type, ZINC ION | | 著者 | Lapi, M, Chaves-Sanjuan, A, Gourlay, L.J, Tiberi, M, Polentarutti, M, Demitri, N, Bais, G, Nardini, M. | | 登録日 | 2022-01-07 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Nuclear factor one X - NFIX in P41212
To Be Published
|
|
7QQD
 
 | | Nuclear factor one X - NFIX in P21 | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NFI binding site (forward), NFI binding site (reverse), ... | | 著者 | Lapi, M, Chaves-Sanjuan, A, Gourlay, L.J, Tiberi, M, Polentarutti, M, Demitri, N, Bais, G, Nardini, M. | | 登録日 | 2022-01-07 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Nuclear factor one X - NFIX in P41212
To Be Published
|
|
6YSB
 
 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) apo form | | 分子名称: | 2-alkenal reductase (NADP(+)-dependent)-like, SULFATE ION | | 著者 | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | 登録日 | 2020-04-21 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
6YUX
 
 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) ternary complex | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, ... | | 著者 | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | 登録日 | 2020-04-27 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
6YTZ
 
 | | Crystal structure of Malus domestica Double Bond Reductase (MdDBR) in complex with NADPH | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, Double Bond Reductase, ... | | 著者 | Caliandro, R, Polsinelli, I, Demitri, N, Benini, S. | | 登録日 | 2020-04-25 | | 公開日 | 2021-02-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | The structural and functional characterization of Malus domestica double bond reductase MdDBR provides insights towards the identification of its substrates.
Int.J.Biol.Macromol., 171, 2021
|
|
6FRW
 
 | | X-ray structure of the levansucrase from Erwinia tasmaniensis | | 分子名称: | GLYCEROL, Levansucrase (Beta-D-fructofuranosyl transferase), ZINC ION | | 著者 | Polsinelli, I, Salomone-Stagni, M, Caliandro, R, Demitri, N, Benini, S. | | 登録日 | 2018-02-16 | | 公開日 | 2019-02-06 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Comparison of the Levansucrase from the epiphyte Erwinia tasmaniensis vs its homologue from the phytopathogen Erwinia amylovora.
Int. J. Biol. Macromol., 127, 2019
|
|
5O7O
 
 | | The crystal structure of DfoC, the desferrioxamine biosynthetic pathway acetyltransferase/Non-Ribosomal Peptide Synthetase (NRPS)-Independent Siderophore (NIS) from the fire blight disease pathogen Erwinia amylovora | | 分子名称: | Desferrioxamine siderophore biosynthesis protein dfoC | | 著者 | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | 登録日 | 2017-06-09 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
6H7G
 
 | | Crystal structure of redox-sensitive phosphoribulokinase (PRK) from the green algae Chlamydomonas reinhardtii | | 分子名称: | Phosphoribulokinase, chloroplastic, SULFATE ION | | 著者 | Fermani, S, Sparla, F, Gurrieri, L, Demitri, N, Polentarutti, M, Falini, G, Trost, P, Lemaire, S.D. | | 登録日 | 2018-07-31 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | ArabidopsisandChlamydomonasphosphoribulokinase crystal structures complete the redox structural proteome of the Calvin-Benson cycle.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6H0U
 
 | | Glycogen synthase kinase-3 beta (GSK3) complex with a covalent [1,2,4]triazolo[1,5-a][1,3,5]triazine inhibitor | | 分子名称: | (2~{R})-3-[7-azanyl-5-(cyclohexylamino)-[1,2,4]triazolo[1,5-a][1,3,5]triazin-2-yl]-2-cyano-propanamide, CHLORIDE ION, GLYCEROL, ... | | 著者 | Marcovich, I, Demitri, N, De Zorzi, R, Storici, P. | | 登録日 | 2018-07-10 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Triazolotriazine-Based Dual GSK-3 beta /CK-1 delta Ligand as a Potential Neuroprotective Agent Presenting Two Different Mechanisms of Enzymatic Inhibition.
Chemmedchem, 14, 2019
|
|
5O5C
 
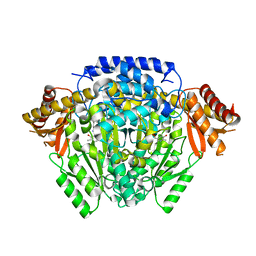 | | The crystal structure of DfoJ, the desferrioxamine biosynthetic pathway lysine decarboxylase from the fire blight disease pathogen Erwinia amylovora | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase involved in desferrioxamine biosynthesis | | 著者 | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | 登録日 | 2017-06-01 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
5O8R
 
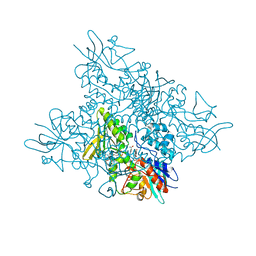 | | The crystal structure of DfoA bound to FAD and NADP; the desferrioxamine biosynthetic pathway cadaverine monooxygenase from the fire blight disease pathogen Erwinia amylovora | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-lysine 6-monooxygenase involved in desferrioxamine biosynthesis, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | 登録日 | 2017-06-14 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J. Struct. Biol., 202, 2018
|
|
5O8P
 
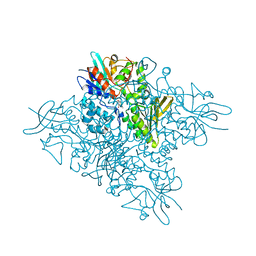 | | The crystal structure of DfoA bound to FAD, the desferrioxamine biosynthetic pathway cadaverine monooxygenase from the fire blight disease pathogen Erwinia amylovora | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, L-lysine 6-monooxygenase involved in desferrioxamine biosynthesis | | 著者 | Salomone-Stagni, M, Bartho, J.D, Polsinelli, I, Bellini, D, Walsh, M.A, Demitri, N, Benini, S. | | 登録日 | 2017-06-14 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | A complete structural characterization of the desferrioxamine E biosynthetic pathway from the fire blight pathogen Erwinia amylovora.
J.Struct.Biol., 202, 2018
|
|
6HK4
 
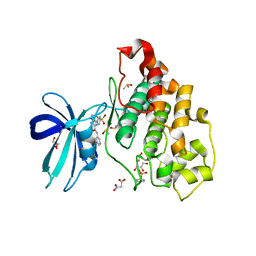 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C22 | | 分子名称: | 3-azanyl-6-(4-morpholin-4-ylsulfonylphenyl)-~{N}-pyridin-3-yl-pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLY-SER-HIS-GLY-HIS-HIS-HIS-HIS-HIS, ... | | 著者 | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | 登録日 | 2018-09-05 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6HK3
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C44 | | 分子名称: | 3-azanyl-~{N}-(2-methoxyphenyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | 登録日 | 2018-09-05 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6HK7
 
 | | Crystal structure of GSK-3B in complex with pyrazine inhibitor C50 | | 分子名称: | 3-azanyl-~{N}-(2-methoxyethyl)-6-[4-(4-methylpiperazin-1-yl)sulfonylphenyl]pyrazine-2-carboxamide, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Piretti, V, Giabbai, B, Demitri, N, Di Martino, R, Tripathi, S.K, Gobbo, D, Decherchi, S, Storici, P, Girotto, S, Cavalli, A. | | 登録日 | 2018-09-05 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Investigating Drug-Target Residence Time in Kinases through Enhanced Sampling Simulations.
J Chem Theory Comput, 15, 2019
|
|
6HQZ
 
 | | Crystal structure of the type III effector protein AvrRpt2 from Erwinia amylovora, a C70 family cysteine protease | | 分子名称: | AvrRpt2 | | 著者 | Bartho, J.D, Demitri, N, Wuerges, J, Benini, S. | | 登録日 | 2018-09-25 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of Erwinia amylovora AvrRpt2 provides insight into protein maturation and induced resistance to fire blight by Malus × robusta 5.
J.Struct.Biol., 206, 2019
|
|
3TOH
 
 | | HIV-1 Protease - Epoxydic Inhibitor Complex (pH 9 - Orthorombic Crystal form P212121) | | 分子名称: | (S)-N-((2S,3S,4R,5R)-4-amino-3,5-dihydroxy-1,6-diphenylhexan-2-yl)-3-methyl-2-(2-phenoxyacetamido)butanamide, DIMETHYL SULFOXIDE, Gag-Pol polyprotein | | 著者 | Geremia, S, Olajuyigbe, F.M, Demitri, N. | | 登録日 | 2011-09-05 | | 公開日 | 2012-08-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.116 Å) | | 主引用文献 | Developing HIV-1 Protease Inhibitors through Stereospecific Reactions in Protein Crystals.
Molecules, 21, 2016
|
|
