1WZ4
 
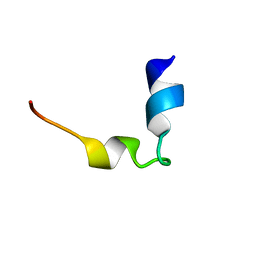 | |
2MPS
 
 | |
5GLJ
 
 | |
6K2K
 
 | | Solution structure of MUL1-RING domain | | 分子名称: | Mitochondrial ubiquitin ligase activator of NFKB 1, ZINC ION | | 著者 | Lee, M.S, Lee, M.K, Ryu, K.S, Chi, S.W. | | 登録日 | 2019-05-14 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of MUL1-RING domain and its interaction with p53 transactivation domain.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
2XD8
 
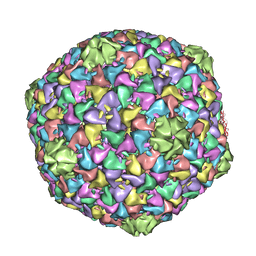 | | Capsid structure of the infectious Prochlorococcus Cyanophage P-SSP7 | | 分子名称: | T7-LIKE CAPSID PROTEIN | | 著者 | Liu, X, Zhang, Q, Murata, K, Baker, M.L, Sullivan, M.B, Fu, C, Dougherty, M, Schmid, M.F, Osburne, M.S, Chisholm, S.W, Chiu, W. | | 登録日 | 2010-04-30 | | 公開日 | 2010-06-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural Changes in a Marine Podovirus Associated with Release of its Genome Into Prochlorococcus
Nat.Struct.Mol.Biol., 17, 2010
|
|
3M7M
 
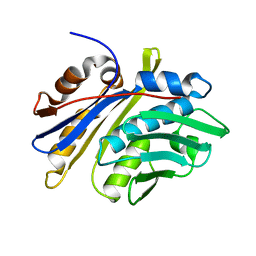 | | Crystal structure of monomeric hsp33 | | 分子名称: | 33 kDa chaperonin | | 著者 | Chi, S.W, Jeong, D.G, Woo, J.R, Park, B.C, Ryu, S.E, Kim, S.J. | | 登録日 | 2010-03-16 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of monomeric hsp33
To be Published
|
|
1COK
 
 | |
1KU0
 
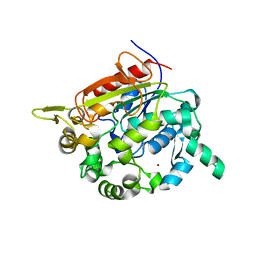 | | Structure of the Bacillus stearothermophilus L1 lipase | | 分子名称: | CALCIUM ION, L1 lipase, ZINC ION | | 著者 | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | 登録日 | 2002-01-18 | | 公開日 | 2002-08-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
7BOL
 
 | |
5GTJ
 
 | |
1I7F
 
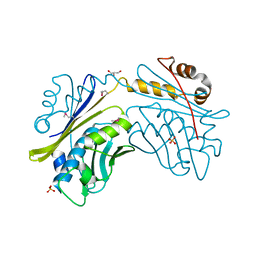 | | CRYSTAL STRUCTURE OF THE HSP33 DOMAIN WITH CONSTITUTIVE CHAPERONE ACTIVITY | | 分子名称: | GLYCEROL, HEAT SHOCK PROTEIN 33, SULFATE ION | | 著者 | Kim, S.-J, Jeong, D.-G, Chi, S.-W, Lee, J.-S, Ryu, S.-E. | | 登録日 | 2001-03-09 | | 公開日 | 2001-05-09 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity
Nat.Struct.Biol., 8, 2001
|
|
2E0T
 
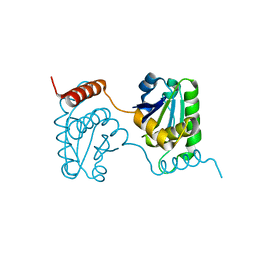 | | Crystal structure of catalytic domain of dual specificity phosphatase 26, MS0830 from Homo sapiens | | 分子名称: | Dual specificity phosphatase 26 | | 著者 | Xie, Y, Kishishita, S, Murayama, K, Hori-Takemoto, C, Chen, L, Liu, Z.J, Wang, B.C, Shirozu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-10-13 | | 公開日 | 2007-10-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | High-resolution crystal structure of the catalytic domain of human dual-specificity phosphatase 26.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6M2D
 
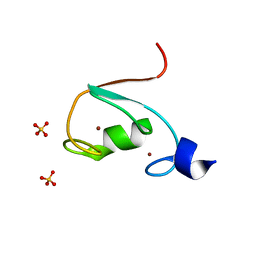 | | MUL1-RING domain | | 分子名称: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | 著者 | Lee, S.O, Ryu, K.S, Chi, S.-W. | | 登録日 | 2020-02-27 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.795 Å) | | 主引用文献 | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
6M2C
 
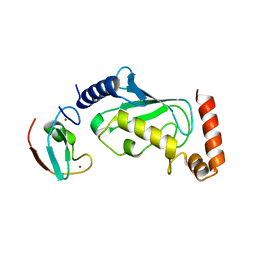 | |
3HJZ
 
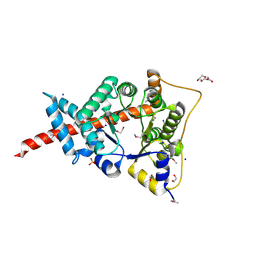 | | The structure of an aldolase from Prochlorococcus marinus | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Singer, A.U, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-05-22 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6VHJ
 
 | |
6VLJ
 
 | |
6VJQ
 
 | |
3FCH
 
 | |
3F56
 
 | |
7JVF
 
 | |
7JU9
 
 | |
6KLY
 
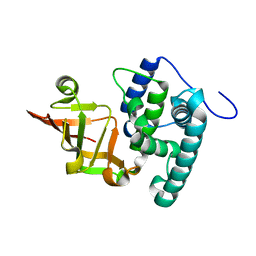 | | Crystal structure of the type III effector XopAI from Xanthomonas axonopodis pv. citri in space group P43212 | | 分子名称: | Type III effector XopAI | | 著者 | Liu, J.-H, Wu, J.E, Lin, H, Chiu, S.W, Yang, J.Y. | | 登録日 | 2019-07-30 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal Structure-Based Exploration of Arginine-Containing Peptide Binding in the ADP-Ribosyltransferase Domain of the Type III Effector XopAI Protein.
Int J Mol Sci, 20, 2019
|
|
