5J0Z
 
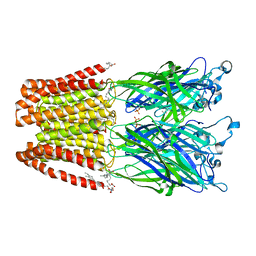 | | Crystal structure of GLIC in complex with DHA | | 分子名称: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | 著者 | Basak, S, Schmandt, N, Chakrapani, S. | | 登録日 | 2016-03-28 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel.
Elife, 6, 2017
|
|
4IX9
 
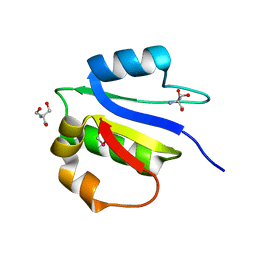 | | Crystal structure of subunit F of V-ATPase from S. cerevisiae | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, V-type proton ATPase subunit F | | 著者 | Basak, S, Balakrishna, A.M, Manimekalai, M.S.S, Gruber, G. | | 登録日 | 2013-01-24 | | 公開日 | 2013-03-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal and NMR structures give insights into the role and dynamics of subunit F of the eukaryotic V-ATPase from Saccharomyces cerevisiae
J.Biol.Chem., 288, 2013
|
|
6NP0
 
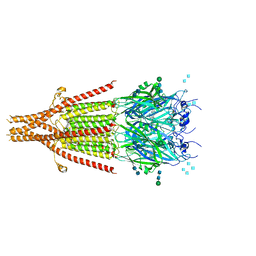 | | Cryo-EM structure of 5HT3A receptor in presence of granisetron | | 分子名称: | 1-methyl-N-[(1R,5S)-9-methyl-9-azabicyclo[3.3.1]nonan-3-yl]indazole-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Basak, S, Chakrapani, S. | | 登録日 | 2019-01-17 | | 公開日 | 2019-07-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | Molecular mechanism of setron-mediated inhibition of full-length 5-HT3Areceptor.
Nat Commun, 10, 2019
|
|
6W1M
 
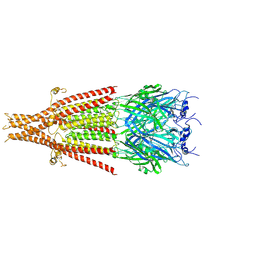 | | Cryo-EM structure of 5HT3A receptor in presence of Ondansetron | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | 著者 | Basak, S, Chakrapani, S. | | 登録日 | 2020-03-04 | | 公開日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | High-resolution structures of multiple 5-HT 3A R-setron complexes reveal a novel mechanism of competitive inhibition.
Elife, 9, 2020
|
|
6W1Y
 
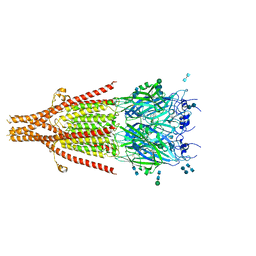 | | Cryo-EM structure of 5HT3A receptor in presence of Palonosetron | | 分子名称: | (3~{a}~{S})-2-[(3~{S})-1-azabicyclo[2.2.2]octan-3-yl]-3~{a},4,5,6-tetrahydro-3~{H}-benzo[de]isoquinolin-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Basak, S, Chakrapani, S. | | 登録日 | 2020-03-04 | | 公開日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | High-resolution structures of multiple 5-HT 3A R-setron complexes reveal a novel mechanism of competitive inhibition.
Elife, 9, 2020
|
|
6W1J
 
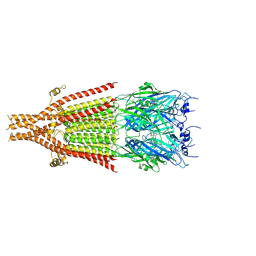 | | Cryo-EM structure of 5HT3A receptor in presence of Alosetron | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(4-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | 著者 | Basak, S, Chakrapani, S. | | 登録日 | 2020-03-04 | | 公開日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | High-resolution structures of multiple 5-HT 3A R-setron complexes reveal a novel mechanism of competitive inhibition.
Elife, 9, 2020
|
|
6DG8
 
 | |
6DG7
 
 | |
6BE1
 
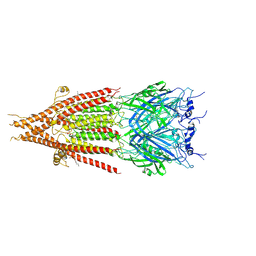 | | Cryo-EM structure of serotonin receptor | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Basak, S, Chakrapani, S. | | 登録日 | 2017-10-24 | | 公開日 | 2018-02-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.31 Å) | | 主引用文献 | Cryo-EM structure of 5-HT
Nat Commun, 9, 2018
|
|
2LX5
 
 | |
8I47
 
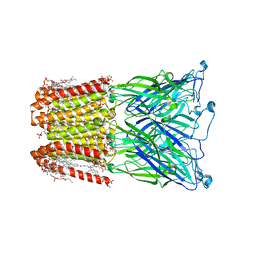 | |
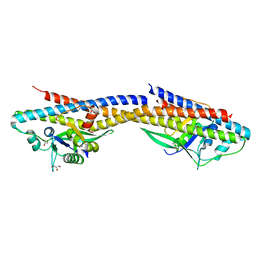
4RND
 
 | |
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-09-13 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8WCQ
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-09-13 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
6B5V
 
 | | Structure of TRPV5 in complex with econazole | | 分子名称: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, Transient receptor potential cation channel subfamily V member 5 | | 著者 | Hughes, T.E.T, Lodowski, D.T, Huynh, K.W, Yazici, A, del Rosario, J, Kapoor, A, Basak, S, Samanta, A, Chakrapani, S, Zhou, Z.H, Filizola, M, Rohacs, T, Han, S, Moiseenkova-Bell, V.Y. | | 登録日 | 2017-09-29 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
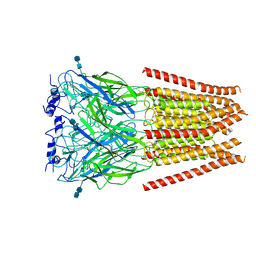
6UBT
 
 | |
6UD3
 
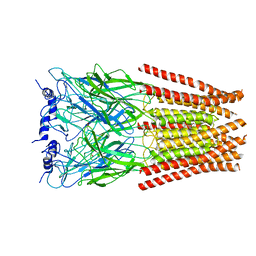 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/PTX-bound open/blocked conformation | | 分子名称: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2019-09-18 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
6VM0
 
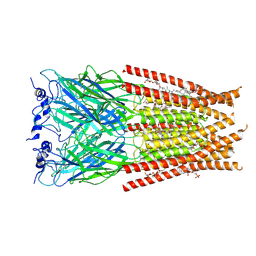 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/IVM-conformation (State-1) | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2020-01-27 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
6VM3
 
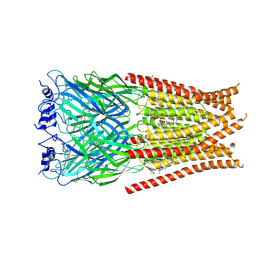 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/IVM-conformation (State-3) | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2020-01-27 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
6VM2
 
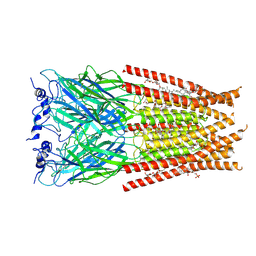 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/IVM-conformation (State-2) | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2020-01-27 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
6UBS
 
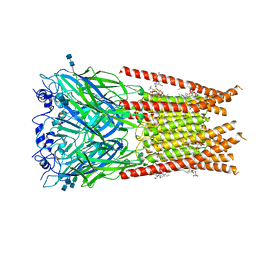 | | Full length Glycine receptor reconstituted in lipid nanodisc in Apo/Resting conformation | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2019-09-12 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-08-12 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
8I42
 
 | |
8I41
 
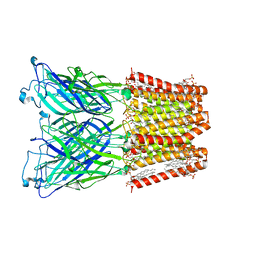 | |
8I48
 
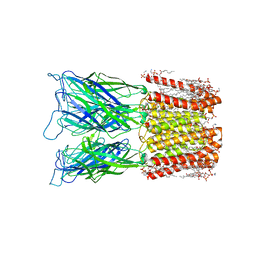 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-01-18 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.74 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8JJ3
 
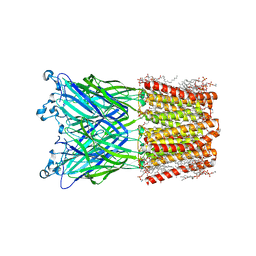 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5 | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | 著者 | Bharambe, N, Li, Z, Basak, S. | | 登録日 | 2023-05-29 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.6476 Å) | | 主引用文献 | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
