7W69
 
 | | Crystal structure of a PSH1 mutant in complex with EDO | | 分子名称: | 1,2-ETHANEDIOL, PSH1 | | 著者 | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | 登録日 | 2021-12-01 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6O
 
 | | Crystal structure of a PSH1 in complex with J1K | | 分子名称: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | 著者 | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | 登録日 | 2021-12-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6Q
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | 分子名称: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | 著者 | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | 登録日 | 2021-12-02 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7CQZ
 
 | | crystal structure of mouse FAM46C (TENT5C) | | 分子名称: | CHLORIDE ION, GLYCEROL, Terminal nucleotidyltransferase 5C | | 著者 | Hu, J.L, Zhang, H, Gao, S. | | 登録日 | 2020-08-12 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.34969759 Å) | | 主引用文献 | Structural and functional characterization of multiple myeloma associated cytoplasmic poly(A) polymerase FAM46C.
Cancer Commun (Lond), 41, 2021
|
|
7CUV
 
 | |
7EEA
 
 | |
7EEQ
 
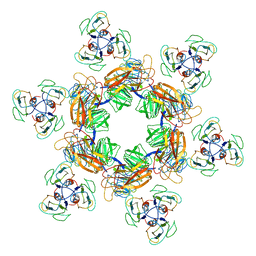 | | Cyanophage Pam1 tail machine | | 分子名称: | Needle head proteins, Tailspike head-binding domain | | 著者 | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | 登録日 | 2021-03-19 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
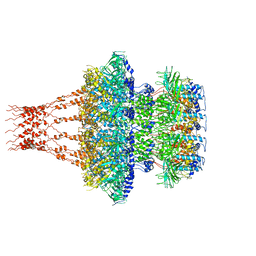
7EEP
 
 | |
7EEL
 
 | |
7E30
 
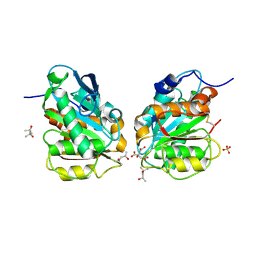 | | Crystal structure of a novel alpha/beta hydrolase in apo form in complex with citrate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRIC ACID, SULFATE ION, ... | | 著者 | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | 登録日 | 2021-02-07 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7E31
 
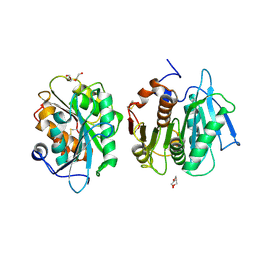 | | Crystal structure of a novel alpha/beta hydrolase mutant in apo form | | 分子名称: | TRIETHYLENE GLYCOL, alpha/beta hydrolase | | 著者 | Gao, J, Han, X, Zheng, Y.Y, Liu, W.D. | | 登録日 | 2021-02-07 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7F2E
 
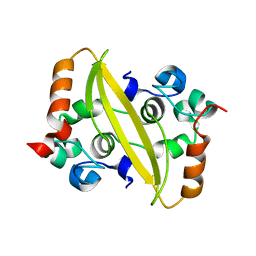 | |
7CMO
 
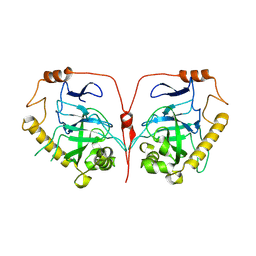 | |
7FJ2
 
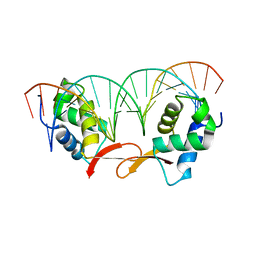 | |
7C02
 
 | | Crystal structure of dimeric MERS-CoV receptor binding domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Dai, L, Qi, J, Gao, G.F. | | 登録日 | 2020-04-30 | | 公開日 | 2020-07-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS.
Cell, 182, 2020
|
|
7BYI
 
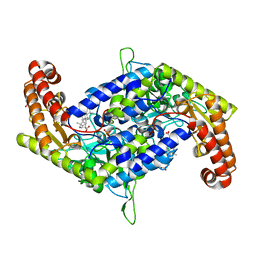 | | Structure of SHMT2 in complex with CBX | | 分子名称: | CARBENOXOLONE, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | 著者 | Li, L, Chen, Y, Lu, D, Zhang, C, Su, D. | | 登録日 | 2020-04-23 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Structure of SHMT2 with CBX
To Be Published
|
|
5ZNK
 
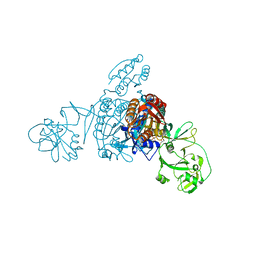 | | Crystal structure of a bacterial ProRS with ligands | | 分子名称: | 7-chloro-6-fluoro-3-{2-oxo-3-[(2S)-piperidin-2-yl]propyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Cheng, B, Yu, Y, Zhou, H. | | 登録日 | 2018-04-09 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
5ZNJ
 
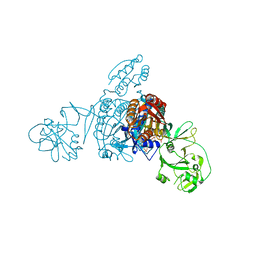 | | Crystal structure of a bacterial ProRS with ligands | | 分子名称: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Cheng, B, Yu, Y, Zhou, H. | | 登録日 | 2018-04-09 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
6XG6
 
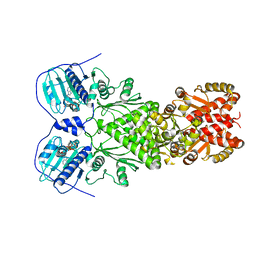 | | Full-length human mitochondrial Hsp90 (TRAP1) with ADP-BeF3 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Heat shock protein 75 kDa, ... | | 著者 | Liu, Y.X, Wang, F, Agard, D.A. | | 登録日 | 2020-06-17 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | General and robust covalently linked graphene oxide affinity grids for high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8EUF
 
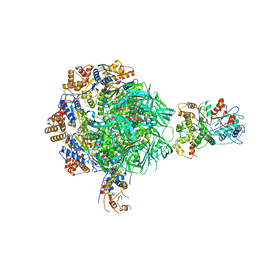 | | Class2 of the INO80-Nucleosome complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | 著者 | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | 登録日 | 2022-10-18 | | 公開日 | 2023-07-12 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUJ
 
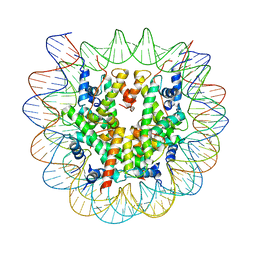 | | Class2 of the INO80-Nucleosome complex | | 分子名称: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | 登録日 | 2022-10-18 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.36 Å) | | 主引用文献 | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETT
 
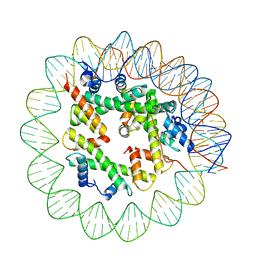 | | Class1 of the INO80-Hexasome complex | | 分子名称: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | 登録日 | 2022-10-17 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (6.68 Å) | | 主引用文献 | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETV
 
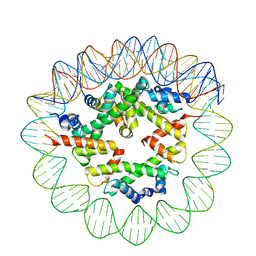 | | Class2 of the INO80-Hexasome complex | | 分子名称: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | 登録日 | 2022-10-17 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EU2
 
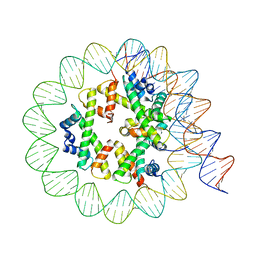 | | Class3 of the INO80-Hexasome complex | | 分子名称: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | 登録日 | 2022-10-18 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUE
 
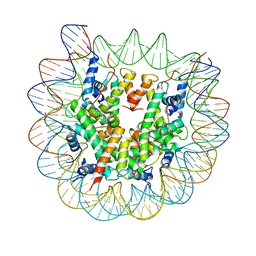 | | Class1 of the INO80-Nucleosome complex | | 分子名称: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | 著者 | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | 登録日 | 2022-10-18 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | ELECTRON MICROSCOPY (3.48 Å) | | 主引用文献 | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
