2MHM
 
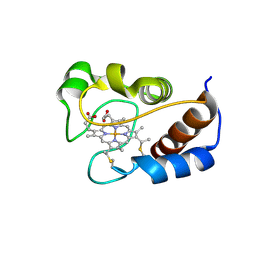 | | Solution structure of cytochrome c Y67H | | 分子名称: | Cytochrome c iso-1, HEME C | | 著者 | Lan, W.X, Wang, Z.H, Yang, Z.Z, Ying, T.L, Wu, H.M, Tan, X.S, Cao, C.Y, Huang, Z.X. | | 登録日 | 2013-11-26 | | 公開日 | 2014-10-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for Cytochrome c Y67H Mutant to Function as a Peroxidase
Plos One, 9, 2014
|
|
5EMY
 
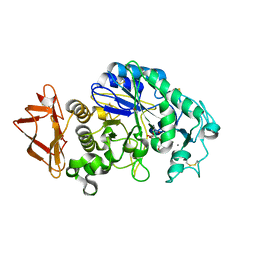 | | Human Pancreatic Alpha-Amylase in complex with the mechanism based inactivator glucosyl epi-cyclophellitol | | 分子名称: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Caner, S, Brayer, G.D. | | 登録日 | 2015-11-06 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.231 Å) | | 主引用文献 | Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic alpha-amylase.
Febs Lett., 590, 2016
|
|
3NVK
 
 | | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle | | 分子名称: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | 著者 | Xue, S, Wang, R, Li, H. | | 登録日 | 2010-07-08 | | 公開日 | 2011-07-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (3.209 Å) | | 主引用文献 | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
4Q2K
 
 | | Bovine alpha chymotrypsin bound to a cyclic peptide inhibitor, 5b | | 分子名称: | (11S)-4,9-dioxo-N-[(2S)-1-oxo-3-phenylpropan-2-yl]-17,22-dioxa-10,30-diazatetracyclo[21.2.2.2~13,16~.1~5,8~]triaconta-1(25),5,7,13,15,23,26,28-octaene-11-carboxamide, Chymotrypsinogen A | | 著者 | Chan, H.Y, Bruning, J.B, Abell, A.D. | | 登録日 | 2014-04-09 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Macrocyclic protease inhibitors with reduced peptide character.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3NVM
 
 | |
7DZ9
 
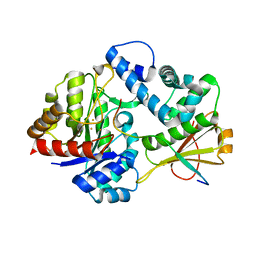 | | MbnABC complex | | 分子名称: | FE (III) ION, MbnA, MbnB, ... | | 著者 | Chao, D, Dan, Z, Yijun, G, Wei, C. | | 登録日 | 2021-01-25 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
3NVI
 
 | |
5XZ3
 
 | | The X-ray structure of Apis mellifera PGRP-SA | | 分子名称: | Peptidoglycan-recognition protein, SULFATE ION | | 著者 | Liu, Y.J, Huang, J.X, Zhao, X.M, An, J.D. | | 登録日 | 2017-07-11 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of peptidoglycan recognition protein SA in Apis mellifera (Hymenoptera: Apidae).
Protein Sci., 27, 2018
|
|
7W0S
 
 | | TRIM7 in complex with C-terminal peptide of 2C | | 分子名称: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase TRIM7, GLYCEROL, ... | | 著者 | Zhang, H, Liang, X, Li, X.Z. | | 登録日 | 2021-11-18 | | 公開日 | 2022-08-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | A C-terminal glutamine recognition mechanism revealed by E3 ligase TRIM7 structures.
Nat.Chem.Biol., 18, 2022
|
|
7W0Q
 
 | |
7W0T
 
 | |
3OZI
 
 | | Crystal structure of the TIR domain from the flax disease resistance protein L6 | | 分子名称: | COBALT (II) ION, L6tr | | 著者 | Ve, T, Bernoux, M, Williams, S, Valkov, E, Warren, C, Hatters, D, Ellis, J.G, Dodds, P.N, Kobe, B. | | 登録日 | 2010-09-25 | | 公開日 | 2011-04-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation.
Cell Host Microbe, 9, 2011
|
|
5H06
 
 | | Crystal structure of AmyP in complex with maltose | | 分子名称: | AmyP, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | He, C, Liu, Y. | | 登録日 | 2016-10-03 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of a raw-starch-degrading bacterial alpha-amylase belonging to subfamily 37 of the glycoside hydrolase family GH13
Sci Rep, 7, 2017
|
|
3P8B
 
 | |
5UJ3
 
 | | Crystal structure of the KPC-2 beta-lactamase complexed with hydrolyzed cefotaxime | | 分子名称: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | 著者 | Pemberton, O.A, Chen, Y. | | 登録日 | 2017-01-16 | | 公開日 | 2017-04-26 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Molecular Basis of Substrate Recognition and Product Release by the Klebsiella pneumoniae Carbapenemase (KPC-2).
J. Med. Chem., 60, 2017
|
|
4RF3
 
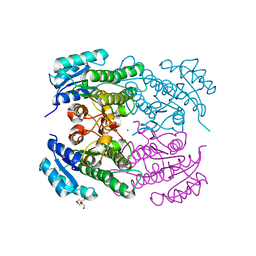 | | Crystal Structure of ketoreductase from Lactobacillus kefir, mutant A94F | | 分子名称: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | 著者 | Tang, Y, Tibrewal, N, Cascio, D. | | 登録日 | 2014-09-24 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.694 Å) | | 主引用文献 | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RF5
 
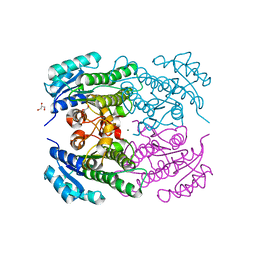 | | Crystal structure of ketoreductase from Lactobacillus kefir, E145S mutant | | 分子名称: | GLYCEROL, MAGNESIUM ION, NADPH dependent R-specific alcohol dehydrogenase | | 著者 | Tang, Y, Tibrewal, N, Cascio, D. | | 登録日 | 2014-09-24 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.596 Å) | | 主引用文献 | Origins of stereoselectivity in evolved ketoreductases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7XPQ
 
 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | 分子名称: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | 著者 | Chen, Y.H, Wang, X.C, Zhang, C.R. | | 登録日 | 2022-05-05 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPP
 
 | |
6IF4
 
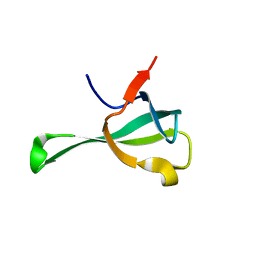 | | Crystal structure of Tbtudor | | 分子名称: | Histone acetyltransferase | | 著者 | Gao, J, Ye, K, Diwu, Y, Liao, S, Tu, X. | | 登録日 | 2018-09-18 | | 公開日 | 2019-09-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.934 Å) | | 主引用文献 | Crystal structure of TbEsa1 presumed Tudor domain from Trypanosoma brucei.
J.Struct.Biol., 209, 2020
|
|
3NMU
 
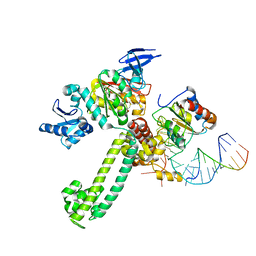 | | Crystal Structure of substrate-bound halfmer box C/D RNP | | 分子名称: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | 著者 | Li, H, Xue, S, Wang, R. | | 登録日 | 2010-06-22 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.729 Å) | | 主引用文献 | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3NCA
 
 | | X-ray structure of ketohexokinase in complex with a thieno pyridinol compound | | 分子名称: | Ketohexokinase, SULFATE ION, thieno[3,2-b]pyridin-7-ol | | 著者 | Abad, M.C, Gibbs, A.C, Spurlino, J.C. | | 登録日 | 2010-06-04 | | 公開日 | 2010-12-22 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.602 Å) | | 主引用文献 | Electron density guided fragment-based lead discovery of ketohexokinase inhibitors.
J.Med.Chem., 53, 2010
|
|
5G5J
 
 | |
7F29
 
 | |
