3BPX
 
 | | Crystal Structure of MarR | | 分子名称: | 2-HYDROXYBENZOIC ACID, SODIUM ION, Transcriptional regulator | | 著者 | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | 登録日 | 2007-12-19 | | 公開日 | 2008-05-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
5AD3
 
 | | Bivalent binding to BET bromodomains | | 分子名称: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | 著者 | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | 登録日 | 2015-08-19 | | 公開日 | 2016-09-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
1D4L
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | 分子名称: | (10S,13S,1'R)-13-[1'-HYDROXY-2'-(N-P-AMINOBENZENESULFONYL-1''-AMINO-3''-METHYLBUTYL)ETHYL]-8,11-DIOXO-10-ISOPROPYL-2-OXA-9,12-DIAZABICYCLO [13.2.2]NONADECA-15,17,18-TRIENE, HIV-1 PROTEASE, SULFATE ION | | 著者 | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | 登録日 | 1999-10-04 | | 公開日 | 2000-10-11 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
5Z5Q
 
 | | Nukacin ISK-1 in active state | | 分子名称: | Lantibiotic nukacin | | 著者 | Kohda, D, Fujinami, D. | | 登録日 | 2018-01-19 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-07-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
7UWY
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 29_bp_sh3 | | 分子名称: | De novo designed small beta-barrel protein 29_bp_sh3 | | 著者 | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | 登録日 | 2022-05-04 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1V2T
 
 | | Trypsin inhibitor in complex with bovine trypsin variant X(SSFI.Glu)bT.B4 | | 分子名称: | CALCIUM ION, METHYL N-[(4-METHYLPHENYL)SULFONYL]GLYCYL-3-[AMINO(IMINO)METHYL]-D-PHENYLALANINATE, SULFATE ION, ... | | 著者 | Rauh, D, Klebe, G, Stubbs, M.T. | | 登録日 | 2003-10-17 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Understanding protein-ligand interactions: the price of protein flexibility
J.Mol.Biol., 335, 2004
|
|
7UWZ
 
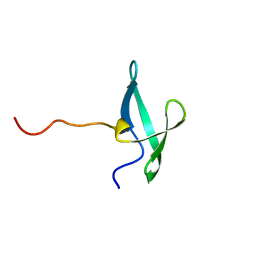 | | NMR solution structure of the De novo designed small beta-barrel protein 33_bp_sh3 | | 分子名称: | De novo designed small beta-barrel protein 33_bp_sh3 | | 著者 | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | 登録日 | 2022-05-04 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8OW2
 
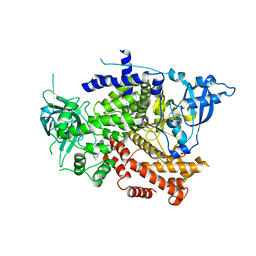 | | Crystal structure of the p110alpha catalytic subunit from homo sapiens in complex with activator 1938 | | 分子名称: | 1-[7-[[2-[[4-(4-ethylpiperazin-1-yl)phenyl]amino]pyridin-4-yl]amino]-2,3-dihydroindol-1-yl]ethanone, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Gong, G.Q, Bellini, D, Vanhaesebroeck, B, Williams, R.L. | | 登録日 | 2023-04-26 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Nature, 618, 2023
|
|
1V2V
 
 | | Benzamidine in complex with bovine trypsin variant X(SSAI)bT.C1 | | 分子名称: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | 著者 | Rauh, D, Klebe, G, Stubbs, M.T. | | 登録日 | 2003-10-17 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Understanding protein-ligand interactions: the price of protein flexibility
J.Mol.Biol., 335, 2004
|
|
1V2N
 
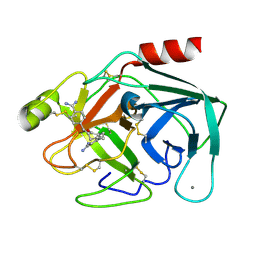 | | Potent factor XA inhibitor in complex with bovine trypsin variant X(99/175/190)bT | | 分子名称: | 2,7-BIS-(4-AMIDINOBENZYLIDENE)-CYCLOHEPTAN-1-ONE, CALCIUM ION, Trypsin | | 著者 | Rauh, D, Klebe, G, Stubbs, M.T. | | 登録日 | 2003-10-17 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Understanding protein-ligand interactions: the price of protein flexibility
J.Mol.Biol., 335, 2004
|
|
3BD9
 
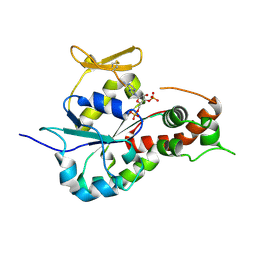 | | human 3-O-sulfotransferase isoform 5 with bound PAP | | 分子名称: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | 著者 | Xu, D, Moon, A.F, Song, D, Liu, J, Pedersen, L.C. | | 登録日 | 2007-11-14 | | 公開日 | 2008-01-29 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Engineering sulfotransferases to modify heparan sulfate.
Nat.Chem.Biol., 4, 2008
|
|
5MST
 
 | |
3N5G
 
 | | Crystal Structure of histidine-tagged human thymidylate synthase | | 分子名称: | SULFATE ION, Thymidylate synthase | | 著者 | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | 登録日 | 2010-05-25 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3N5E
 
 | | Crystal Structure of human thymidylate synthase bound to a peptide inhibitor | | 分子名称: | SULFATE ION, Synthetic peptide LR, Thymidylate synthase | | 著者 | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | 登録日 | 2010-05-25 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5YXW
 
 | | Crystal structure of the prefusion form of measles virus fusion protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, glycoprotein F1,measles virus fusion protein, ... | | 著者 | Hashiguchi, T, Fukuda, Y, Matsuoka, R, Kuroda, D, Kubota, M, Shirogane, Y, Watanabe, S, Tsumoto, K, Kohda, D, Plemper, R.K, Yanagi, Y. | | 登録日 | 2017-12-07 | | 公開日 | 2018-02-21 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.776 Å) | | 主引用文献 | Structures of the prefusion form of measles virus fusion protein in complex with inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1OY7
 
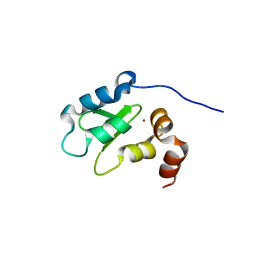 | | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP) | | 分子名称: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, AEVVAVKSE peptide, Baculoviral IAP repeat-containing protein 7, ... | | 著者 | Franklin, M.C, Kadkhodayan, S, Ackerly, H, Alexandru, D, Distefano, M.D, Elliott, L.O, Flygare, J.A, Vucic, D, Deshayes, K, Fairbrother, W.J. | | 登録日 | 2003-04-03 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure and Function Analysis of Peptide Antagonists of Melanoma Inhibitor of Apoptosis (ML-IAP)
Biochemistry, 42, 2003
|
|
6B85
 
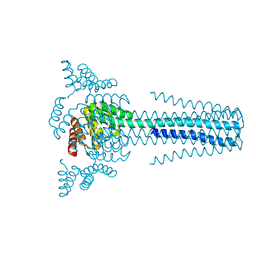 | | Crystal structure of transmembrane protein TMHC4_R | | 分子名称: | TMHC4_R | | 著者 | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | 登録日 | 2017-10-05 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.889 Å) | | 主引用文献 | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6B87
 
 | | Crystal structure of transmembrane protein TMHC2_E | | 分子名称: | TMHC2_E | | 著者 | Lu, P, DiMaio, F, Min, D, Wei, K.Y, Bowie, J, Baker, D. | | 登録日 | 2017-10-05 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.947 Å) | | 主引用文献 | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
7XRP
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with nanobody C5G2 (localized refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5G2 nanobody, Spike protein S1 | | 著者 | Liu, L, Sun, H, Jiang, Y, Liu, X, Zhao, D, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2022-05-11 | | 公開日 | 2022-10-05 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | A potent synthetic nanobody with broad-spectrum activity neutralizes SARS-CoV-2 virus and the Omicron variant BA.1 through a unique binding mode.
J Nanobiotechnology, 20, 2022
|
|
6BBM
 
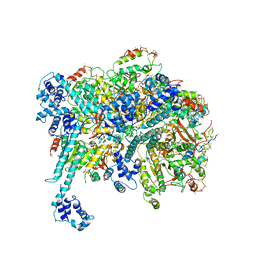 | | Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Replication protein P, Replicative DNA helicase | | 著者 | Chase, J, Catalano, A, Noble, A.J, Eng, E.T, Olinares, P.D.B, Molloy, K, Pakotiprapha, D, Samuels, M, Chain, B, des Georges, A, Jeruzalmi, D. | | 登録日 | 2017-10-18 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Mechanisms of opening and closing of the bacterial replicative helicase.
Elife, 7, 2018
|
|
3BPV
 
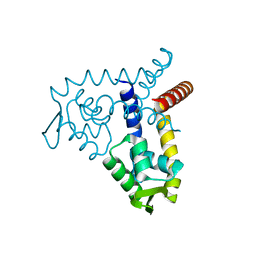 | | Crystal Structure of MarR | | 分子名称: | Transcriptional regulator | | 著者 | Saridakis, V, Shahinas, D, Xu, X, Christendat, D. | | 登録日 | 2007-12-19 | | 公開日 | 2008-05-20 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural insight on the mechanism of regulation of the MarR family of proteins: high-resolution crystal structure of a transcriptional repressor from Methanobacterium thermoautotrophicum.
J.Mol.Biol., 377, 2008
|
|
6C2Z
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-Aminoacrylate Intermediate | | 分子名称: | 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CALCIUM ION, ... | | 著者 | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | 登録日 | 2018-01-09 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
1V2L
 
 | | Benzamidine in complex with bovine trypsin variant X(triple.Glu)bT.D1 | | 分子名称: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | 著者 | Rauh, D, Klebe, G, Stubbs, M.T. | | 登録日 | 2003-10-17 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Understanding protein-ligand interactions: the price of protein flexibility
J.Mol.Biol., 335, 2004
|
|
1V2P
 
 | | Trypsin inhibitor in complex with bovine trypsin variant X(SSYI)bT.A4 | | 分子名称: | CALCIUM ION, METHYL N-[(4-METHYLPHENYL)SULFONYL]GLYCYL-3-[AMINO(IMINO)METHYL]-D-PHENYLALANINATE, SULFATE ION, ... | | 著者 | Rauh, D, Klebe, G, Stubbs, M.T. | | 登録日 | 2003-10-17 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Understanding protein-ligand interactions: the price of protein flexibility
J.Mol.Biol., 335, 2004
|
|
1V2W
 
 | | Trypsin inhibitor in complex with bovine trypsin variant X(SSAI)bT.B4 | | 分子名称: | CALCIUM ION, METHYL N-[(4-METHYLPHENYL)SULFONYL]GLYCYL-3-[AMINO(IMINO)METHYL]-D-PHENYLALANINATE, SULFATE ION, ... | | 著者 | Rauh, D, Klebe, G, Stubbs, M.T. | | 登録日 | 2003-10-17 | | 公開日 | 2004-06-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Understanding protein-ligand interactions: the price of protein flexibility
J.Mol.Biol., 335, 2004
|
|
