5Y14
 
 | | Crystal structure of LP-40/N44 | | 分子名称: | LP-40, N44 | | 著者 | Zhang, X, Wang, X, He, Y. | | 登録日 | 2017-07-19 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.762 Å) | | 主引用文献 | Enfuvirtide (T20)-Based Lipopeptide Is a Potent HIV-1 Cell Fusion Inhibitor: Implications for Viral Entry and Inhibition
J. Virol., 91, 2017
|
|
6LDZ
 
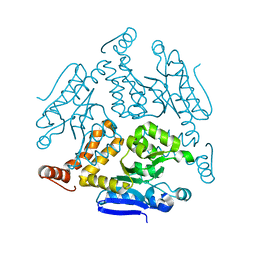 | | Crystal structure of Rv0222 from Mycobacterium tuberculosis | | 分子名称: | Probable enoyl-CoA hydratase EchA1 (Enoyl hydrase) (Unsaturated acyl-CoA hydratase) (Crotonase) | | 著者 | Li, J, Ran, Y.J, Wang, L, Wu, J.H, Ge, B.X, Rao, Z.H. | | 登録日 | 2019-11-23 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Host-mediated ubiquitination of a mycobacterial protein suppresses immunity.
Nature, 577, 2020
|
|
6O1F
 
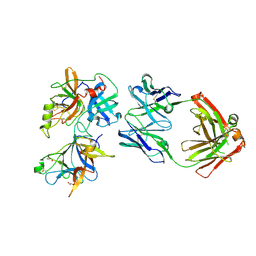 | |
5GMZ
 
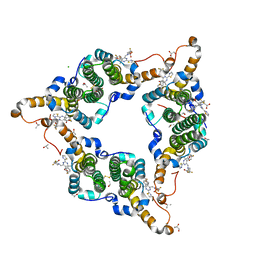 | | Hepatitis B virus core protein Y132A mutant in complex with 4-methyl heteroaryldihydropyrimidine | | 分子名称: | (2S)-4,4-difluoro-1-[[(4S)-4-(4-fluorophenyl)-5-methoxycarbonyl-4-methyl-2-(1,3-thiazol-2-yl)-1H-pyrimidin-6-yl]methyl]pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | 著者 | Xu, Z.H, Zhou, Z. | | 登録日 | 2016-07-18 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Design and Synthesis of Orally Bioavailable 4-Methyl Heteroaryldihydropyrimidine Based Hepatitis B Virus (HBV) Capsid Inhibitors
J.Med.Chem., 59, 2016
|
|
9M7V
 
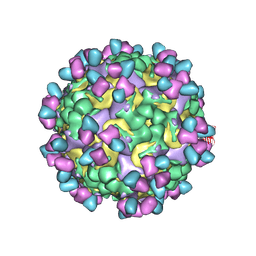 | | Cryo-EM structure of enterovirus A71 mature virion in complex with Fab CT11F9 | | 分子名称: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | 著者 | Jiang, Y, Zhu, R, Zheng, Q, Li, S, Xia, N. | | 登録日 | 2025-03-11 | | 公開日 | 2025-04-23 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Development of In Vitro Potency Methods to Replace In Vivo Tests for Enterovirus 71 Inactivated Vaccine (Human Diploid Cell-Based/Vero Cell-Based).
Vaccines (Basel), 13, 2025
|
|
9L0Q
 
 | |
9JUY
 
 | | X-ray crystal structure of Y16513 in CBP | | 分子名称: | (5~{S})-1-(3-chloranyl-4-methoxy-phenyl)-5-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{S})-2-morpholin-4-ylpropyl]imidazol-2-yl]pyrrolidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE | | 著者 | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | 登録日 | 2024-10-08 | | 公開日 | 2025-03-12 | | 最終更新日 | 2025-06-04 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
9JUT
 
 | | X-ray crystal structure of Y16524 in EP300 | | 分子名称: | (6~{S})-1-(3-chloranyl-4-methoxy-phenyl)-6-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{S})-2-morpholin-4-ylpropyl]imidazol-2-yl]piperidin-2-one, Histone acetyltransferase p300 | | 著者 | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | 登録日 | 2024-10-08 | | 公開日 | 2025-03-12 | | 最終更新日 | 2025-06-04 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
9JUU
 
 | | X-ray crystal structure of Y16515 in CBP | | 分子名称: | (5~{S})-1-(3-chloranyl-4-methoxy-phenyl)-5-[4-(3-methyl-1,2-benzoxazol-5-yl)-1-[(2~{R})-2-morpholin-4-ylpropyl]imidazol-2-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein, ... | | 著者 | Hu, J, Zhang, C, Luo, G, Tang, X, Wu, T, Shen, H, Zhao, X, Wu, X, Smaill, J, Zhang, Y, Xu, Y, Xiang, Q. | | 登録日 | 2024-10-08 | | 公開日 | 2025-03-12 | | 最終更新日 | 2025-06-04 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Discovery of 5-imidazole-3-methylbenz[d]isoxazole derivatives as potent and selective CBP/p300 bromodomain inhibitors for the treatment of acute myeloid leukemia.
Acta Pharmacol.Sin., 46, 2025
|
|
5K9J
 
 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 isolated following H5 immunization. | | 分子名称: | 56.a.09 heavy chain, 56.a.09 light chain, POLYETHYLENE GLYCOL (N=34) | | 著者 | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | 登録日 | 2016-05-31 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.904 Å) | | 主引用文献 | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
8CUH
 
 | |
9J3M
 
 | | ADP/Pi bound Arabidopsis ATP/ADP translocator AtNTT1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | 著者 | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | 登録日 | 2024-08-08 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (2.77 Å) | | 主引用文献 | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3L
 
 | | ATP bound Arabidopsis ATP/ADP translocator AtNTT1 | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1, chloroplastic, ... | | 著者 | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | 登録日 | 2024-08-08 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3N
 
 | | ATP bound Chlamydia pneumoniae ATP/ADP translocator NTT1(Inward open state) | | 分子名称: | 1D10, ADENOSINE-5'-TRIPHOSPHATE, ADP,ATP carrier protein 1 | | 著者 | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | 登録日 | 2024-08-08 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3O
 
 | | Chlamydia pneumoniae ATP/ADP translocator NTT1(Outward open state) | | 分子名称: | 1D10, ADP,ATP carrier protein 1 | | 著者 | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | 登録日 | 2024-08-08 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
9J3J
 
 | | Arabidopsis ATP/ADP translocator AtNTT1 | | 分子名称: | ADP,ATP carrier protein 1, chloroplastic, nanobody: B-C8 | | 著者 | Lin, H.J, Huang, J, Li, T.M, Li, W.J, Su, N.N, Zhang, J.R, Wu, X.D, Fan, M.R. | | 登録日 | 2024-08-08 | | 公開日 | 2025-03-19 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (2.83 Å) | | 主引用文献 | Structure and mechanism of the plastid/parasite ATP/ADP translocator.
Nature, 641, 2025
|
|
6W5S
 
 | | NPC1 structure in GDN micelles at pH 8.0 | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5V
 
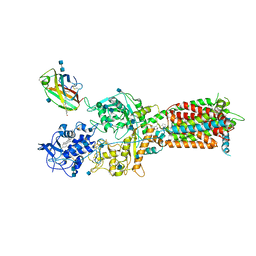 | | NPC1-NPC2 complex structure at pH 5.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5R
 
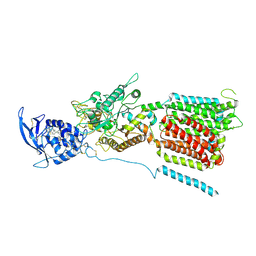 | | NPC1 structure in Nanodisc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5U
 
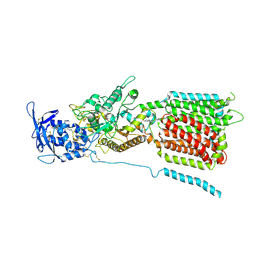 | | NPC1 structure in GDN micelles at pH 5.5, conformation b | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2025-05-21 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6W5T
 
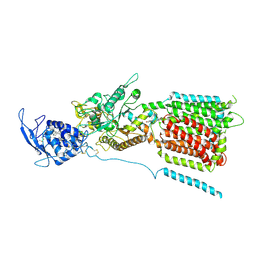 | | NPC1 structure in GDN micelles at pH 5.5, conformation a | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Yan, N, Qian, H.W, Wu, X.L. | | 登録日 | 2020-03-13 | | 公開日 | 2020-06-17 | | 最終更新日 | 2025-05-14 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
3DOR
 
 | | Crystal Structure of mature CPAF | | 分子名称: | Protein CT_858, SULFATE ION | | 著者 | Chai, J, Huang, Z. | | 登録日 | 2008-07-06 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPM
 
 | | Structure of mature CPAF complexed with lactacystin | | 分子名称: | N-acetyl-S-({(2R,3S,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxopyrrolidin-2-yl}carbonyl)cysteine, Protein CT_858 | | 著者 | Chai, J, Huang, Z. | | 登録日 | 2008-07-09 | | 公開日 | 2009-01-13 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DJA
 
 | |
3DPN
 
 | |
