5XXH
 
 | | Crystal Structure Analysis of the CBP | | 分子名称: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | 著者 | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | 登録日 | 2017-07-04 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
3CGL
 
 | |
5K72
 
 | | IRAK4 in complex with Compound 21 | | 分子名称: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]cyclohexane-1,4-diamine | | 著者 | Ferguson, A.D. | | 登録日 | 2016-05-25 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
5K7G
 
 | | IRAK4 in complex with AZ3862 | | 分子名称: | (3~{a}~{S},7~{a}~{R})-1-methyl-5-[4-[[5-(oxan-4-yl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]amino]cyclohexyl]-3,3~{a},4,6,7,7~{a}-hexahydropyrrolo[3,2-c]pyridin-2-one, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | 著者 | Ferguson, A.D. | | 登録日 | 2016-05-26 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Discovery and Optimization of Pyrrolopyrimidine Inhibitors of Interleukin-1 Receptor Associated Kinase 4 (IRAK4) for the Treatment of Mutant MYD88L265P Diffuse Large B-Cell Lymphoma.
J. Med. Chem., 60, 2017
|
|
9H8E
 
 | | Crystal structure of HPK1 T165E/S171E in complex with compound 13 | | 分子名称: | 5-(methylamino)-6-(3-methylimidazo[4,5-c]pyridin-7-yl)-3-[(4-morpholin-4-ylphenyl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | Schimpl, M, Pflug, A. | | 登録日 | 2024-10-29 | | 公開日 | 2025-02-19 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (1.631 Å) | | 主引用文献 | Discovery and Optimization of Pyrazine Carboxamide AZ3246, a Selective HPK1 Inhibitor.
J.Med.Chem., 68, 2025
|
|
9H8D
 
 | | Crystal structure of HPK1 T165E/S171E in complex with compound 6 | | 分子名称: | 6-(1-methylbenzimidazol-4-yl)-3-[(4-morpholin-4-ylphenyl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | Schimpl, M, Pflug, A. | | 登録日 | 2024-10-29 | | 公開日 | 2025-02-19 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Discovery and Optimization of Pyrazine Carboxamide AZ3246, a Selective HPK1 Inhibitor.
J.Med.Chem., 68, 2025
|
|
9H8F
 
 | | Crystal structure of HPK1 T165E/S171E in complex with pyrazine carboxamide inhibitor AZ3246 (compound 24) | | 分子名称: | 1,2-ETHANEDIOL, 5-cyclopropyl-6-(3-methylimidazo[4,5-c]pyridin-7-yl)-3-[[3-methyl-1-[2,2,2-tris(fluoranyl)ethyl]pyrazol-4-yl]amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | 著者 | Schimpl, M, Pflug, A. | | 登録日 | 2024-10-29 | | 公開日 | 2025-02-19 | | 最終更新日 | 2025-03-05 | | 実験手法 | X-RAY DIFFRACTION (1.393 Å) | | 主引用文献 | Discovery and Optimization of Pyrazine Carboxamide AZ3246, a Selective HPK1 Inhibitor.
J.Med.Chem., 68, 2025
|
|
6KZE
 
 | | The crystal structue of PDE10A complexed with 4d | | 分子名称: | 8-[(E)-2-[5-methyl-1-[3-[3-(4-methylpiperazin-1-yl)propoxy]phenyl]benzimidazol-2-yl]ethenyl]quinoline, MAGNESIUM ION, ZINC ION, ... | | 著者 | Yang, Y, Zhang, S, Zhou, Q, Huang, Y.-Y, Guo, L, Luo, H.-B. | | 登録日 | 2019-09-24 | | 公開日 | 2020-09-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.50003481 Å) | | 主引用文献 | Novel Potent and Highly Selective Benzoimidazole-based Phosphodiesterase 10 Inhibitors with Improved Solubility and Pharmacokinetic Properties for the Treatment of Pulmonary Arterial Hypertension
To Be Published
|
|
6ZOS
 
 | | Oestrogen receptor ligand binding domain in complex with compound 18 | | 分子名称: | 6-[(6~{S},8~{R})-7-[(1-fluoranylcyclopropyl)methyl]-8-methyl-2,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-~{N}-[1-(3-fluoranylpropyl)azetidin-3-yl]pyridin-3-amine, Estrogen receptor | | 著者 | Breed, J. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
6ZOR
 
 | | Oestrogen receptor ligand binding domain in complex with compound 28 | | 分子名称: | 6-[(6~{S},8~{R})-8-methyl-7-[2,2,2-tris(fluoranyl)ethyl]-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-~{N}-(1-propylazetidin-3-yl)pyridin-3-amine, Estrogen receptor | | 著者 | Breed, J. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
6ZOQ
 
 | | Oestrogen receptor ligand binding domain in complex with compound 16 | | 分子名称: | Estrogen receptor, ~{N}-[4-[(6~{S},8~{R})-7-[(1-fluoranylcyclopropyl)methyl]-8-methyl-2,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-3-methoxy-phenyl]-1-(3-fluoranylpropyl)azetidin-3-amine | | 著者 | Breed, J. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
7RPV
 
 | |
8HM1
 
 | |
8HM3
 
 | | Complex of PPIase-BfUbb | | 分子名称: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | 著者 | Xu, J.H, Chen, Z, Gao, X. | | 登録日 | 2022-12-02 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8HM2
 
 | |
8HM4
 
 | | Crystal structure of PPIase | | 分子名称: | Peptidylprolyl isomerase | | 著者 | Xu, J.H, Chen, Z, Gao, X. | | 登録日 | 2022-12-02 | | 公開日 | 2023-11-29 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (3.79 Å) | | 主引用文献 | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
7Y99
 
 | | Crystal Structure Analysis of cp2 bound BCLxl | | 分子名称: | Bcl-2-like protein 1, CP2 peptide, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide | | 著者 | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | 登録日 | 2022-06-24 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YAA
 
 | | Crystal structure analysis of cp3 bound BCLxl | | 分子名称: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | 著者 | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | 登録日 | 2022-06-27 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YA5
 
 | | Crystal structure analysis of cp1 bound BCL2/G101V | | 分子名称: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | 著者 | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | 登録日 | 2022-06-27 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y8D
 
 | | Crystal structure of cp1 bound BCLxl | | 分子名称: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Bcl-2-like protein 1, cp1 peptide | | 著者 | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | 登録日 | 2022-06-23 | | 公開日 | 2023-11-15 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
6WLW
 
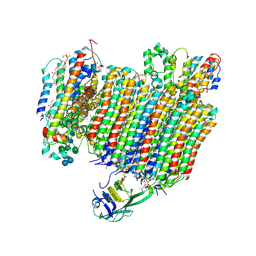 | | The Vo region of human V-ATPase in state 1 (focused refinement) | | 分子名称: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, L, Wu, H, Fu, T.-M. | | 登録日 | 2020-04-20 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WM2
 
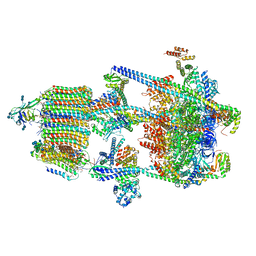 | | Human V-ATPase in state 1 with SidK and ADP | | 分子名称: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, L, Wu, H, Fu, T.M. | | 登録日 | 2020-04-20 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WLZ
 
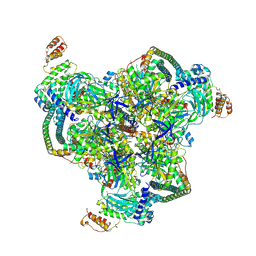 | | The V1 region of human V-ATPase in state 1 (focused refinement) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, SidK, V-type proton ATPase catalytic subunit A, ... | | 著者 | Wang, L, Wu, H, Fu, T.M. | | 登録日 | 2020-04-20 | | 公開日 | 2020-11-11 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
6WM3
 
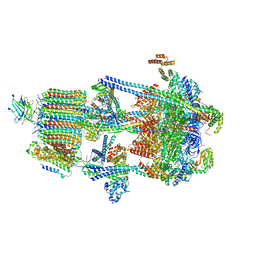 | | Human V-ATPase in state 2 with SidK and ADP | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Renin receptor, ... | | 著者 | Wang, L, Wu, H, Fu, T.M. | | 登録日 | 2020-04-20 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of a Complete Human V-ATPase Reveal Mechanisms of Its Assembly.
Mol.Cell, 80, 2020
|
|
8J6O
 
 | | transport T2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Green fluorescent protein (Fragment),SID1 transmembrane family member 2, ... | | 著者 | Jiang, D.H, Zhang, J.T. | | 登録日 | 2023-04-26 | | 公開日 | 2024-05-01 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
