5GT1
 
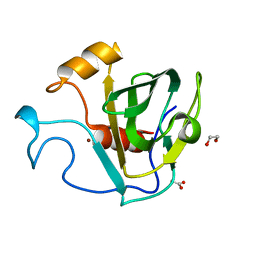 | | crystal structure of cbpa from L. salivarius REN | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Choline binding protein A, ... | | 著者 | Jiang, L, Ren, F. | | 登録日 | 2016-08-18 | | 公開日 | 2017-07-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial Cell Line Requires S-layer Proteins
Sci Rep, 7, 2017
|
|
5HNQ
 
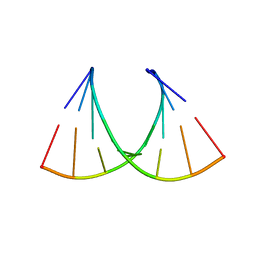 | |
5HN2
 
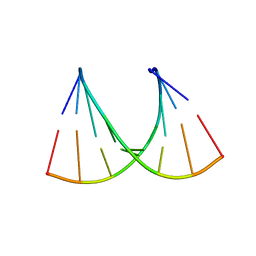 | |
5HNJ
 
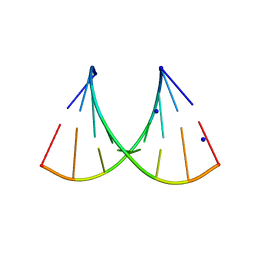 | |
2L8E
 
 | |
5M25
 
 | | Modulation of MLL1 Methyltransferase Activity | | 分子名称: | (2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-~{N}-[(2~{S})-1-[[4-[(~{E})-[4-(hydroxymethyl)phenyl]diazenyl]phenyl]methylamino]-1-oxidanylidene-propan-2-yl]pentanamide, WD repeat-containing protein 5 | | 著者 | Srinivasan, V. | | 登録日 | 2016-10-11 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
5M23
 
 | | Modulation of MLL1 Methyltransferase Activity | | 分子名称: | 4-[(~{E})-[4-[[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-pentanoyl]amino]propanoyl]amino]methyl]phenyl]diazenyl]-~{N}-[(2~{S})-3-methyl-1-oxidanylidene-butan-2-yl]benzamide, WD repeat-containing protein 5 | | 著者 | Srinivasan, V. | | 登録日 | 2016-10-11 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | 分子名称: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | 著者 | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | 登録日 | 2016-12-19 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (4.17 Å) | | 主引用文献 | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5MPS
 
 | | Structure of a spliceosome remodeled for exon ligation | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | 登録日 | 2016-12-18 | | 公開日 | 2017-01-18 | | 最終更新日 | 2020-10-07 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
6UAI
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with YSAM peptide | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Toth, E.A, Bryan, P.N, Orban, J. | | 登録日 | 2019-09-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UAO
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with the peptide EEYSAM | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptide EEYSAM, ... | | 著者 | Toth, E.A, Bryan, P.N, Orban, J. | | 登録日 | 2019-09-11 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
3VF9
 
 | |
3V8T
 
 | |
3VF8
 
 | |
3V8W
 
 | |
7MU5
 
 | | Human DCTPP1 bound to Triptolide | | 分子名称: | MAGNESIUM ION, dCTP pyrophosphatase 1, triptolide | | 著者 | Hauk, G, Berger, J.M. | | 登録日 | 2021-05-14 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Triptolide sensitizes cancer cells to nucleoside DNA methyltransferase inhibitors through inhibition of DCTPP1 mediated cell-intrinsic resistance
To Be Published
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | 分子名称: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2J
 
 | | Crystal structure of nanobody Nb70 with SARS-CoV RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nb70, Spike protein S1 | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2L
 
 | | Crystal structure of nanobody 3-2A2-4 with SARS-CoV-2 RBD | | 分子名称: | Nanobody 3-2A2-4, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7X2K
 
 | | Crystal structure of nanobody Nb70 with antibody 1F11 fab and SARS-CoV-2 RBD | | 分子名称: | 1F11-H, 1F11-L, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | 登録日 | 2022-02-25 | | 公開日 | 2022-12-21 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
7XGW
 
 | |
8JM9
 
 | |
8JMI
 
 | |
8JMA
 
 | |
8JME
 
 | |
