6CV4
 
 | |
5V00
 
 | | Structure of HutD from Pseudomonas fluorescens SBW25 (Formate condition) | | 分子名称: | FORMIC ACID, GLYCEROL, Uncharacterized protein | | 著者 | Liu, Y, Johnston, J.M, Gerth, M.L, Baker, E.N, Lott, J.S, Rainey, P.B. | | 登録日 | 2017-02-27 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a bicupin protein HutD involved in histidine utilization in Pseudomonas.
Proteins, 85, 2017
|
|
4QQG
 
 | | Crystal structure of an N-terminal HTATIP fragment | | 分子名称: | Histone acetyltransferase KAT5, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Wernimont, A.K, Dobrovetsky, E, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2014-06-27 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural and histone binding studies of the chromo barrel domain of TIP60.
FEBS Lett., 592, 2018
|
|
4RGY
 
 | |
6IH5
 
 | | Crystal structure of Phosphite Dehydrogenase mutant I151R/P176E from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | 分子名称: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | 登録日 | 2018-09-28 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.468 Å) | | 主引用文献 | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH4
 
 | |
6IH2
 
 | |
6IH3
 
 | | Crystal structure of Phosphite Dehydrogenase from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine Dinucleotide | | 分子名称: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Song, X, Feng, Y, Zhao, Z, Liu, Y. | | 登録日 | 2018-09-28 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.942 Å) | | 主引用文献 | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH6
 
 | | Phosphite Dehydrogenase mutant I151R/P176R/M207A from Ralstonia sp. 4506 in complex with non-natural cofactor Nicotinamide Cytosine dinucleotide | | 分子名称: | Phosphite dehydrogenase, [[(2S,3S,4R,5S)-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4R,5S)-5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | 著者 | Song, X, Feng, Y, Liu, Y, Zhao, Z. | | 登録日 | 2018-09-28 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.491 Å) | | 主引用文献 | Structural Insights into Phosphite Dehydrogenase Variants Favoring a Non-natural Redox Cofactor
Acs Catalysis, 9, 2019
|
|
6IH8
 
 | |
4RZI
 
 | | Crystal structure of PhaB from Synechocystis sp. PCC 6803 | | 分子名称: | 3-ketoacyl-acyl carrier protein reductase | | 著者 | Xue, S, Liu, Y. | | 登録日 | 2014-12-22 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.891 Å) | | 主引用文献 | Structure-directed construction of a high-performance version of the enzyme FabG from the photosynthetic microorganism Synechocystis sp. PCC 6803.
Febs Lett., 589, 2015
|
|
1LXT
 
 | | STRUCTURE OF PHOSPHOTRANSFERASE PHOSPHOGLUCOMUTASE FROM RABBIT | | 分子名称: | CADMIUM ION, PHOSPHOGLUCOMUTASE (DEPHOSPHO FORM), SULFATE ION | | 著者 | Ray Junior, W.J, Baranidharan, S, Liu, Y. | | 登録日 | 1996-07-28 | | 公開日 | 1997-02-12 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of rabbit muscle phosphoglucomutase refined at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
8X3R
 
 | |
8X3S
 
 | | Crystal structure of human WDR5 in complex with PTEN | | 分子名称: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, WD repeat-containing protein 5 | | 著者 | Liu, Y, Huang, X, Shang, X. | | 登録日 | 2023-11-14 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
6V2S
 
 | | Crystal Structure of chromodomain of MPP8 in complex with inhibitor UNC3866 | | 分子名称: | M-phase phosphoprotein 8, UNC3866, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2019-11-25 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2R
 
 | | Crystal Structure of chromodomain of CBX7 mutant V13A in complex with inhibitor UNC3866 | | 分子名称: | Chromobox protein homolog 7, UNC3866, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2019-11-25 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6V2D
 
 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | 分子名称: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2019-11-22 | | 公開日 | 2019-12-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6GRC
 
 | | eukaryotic junction-resolving enzyme GEN-1 binding with Sodium | | 分子名称: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | 著者 | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | 登録日 | 2018-06-11 | | 公開日 | 2018-09-26 | | 最終更新日 | 2019-02-13 | | 実験手法 | X-RAY DIFFRACTION (2.452 Å) | | 主引用文献 | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
6GRD
 
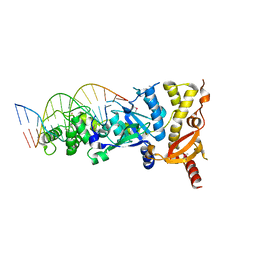 | | eukaryotic junction-resolving enzyme GEN-1 binding with Cesium | | 分子名称: | CESIUM ION, DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), ... | | 著者 | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | 登録日 | 2018-06-11 | | 公開日 | 2018-09-26 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
6GRB
 
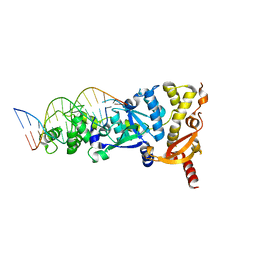 | | eukaryotic junction-resolving enzyme GEN-1 binding with Potassium | | 分子名称: | DNA (5'-D(*TP*AP*CP*CP*CP*AP*CP*CP*AP*CP*CP*GP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*CP*GP*GP*TP*GP*GP*TP*TP*GP*GP*T)-3'), MAGNESIUM ION, ... | | 著者 | Lilley, D.M.J, Liu, Y, Freeman, D.J. | | 登録日 | 2018-06-11 | | 公開日 | 2018-09-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A monovalent ion in the DNA binding interface of the eukaryotic junction-resolving enzyme GEN1.
Nucleic Acids Res., 46, 2018
|
|
8VCN
 
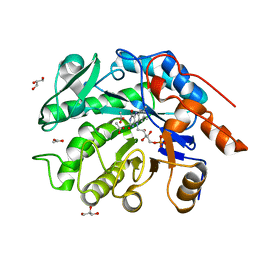 | | GluER mutant - W66F F269Y Q293T F68Y T36E P263L | | 分子名称: | ACETATE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | 著者 | Jeffrey, P.D, Sorigue, D.R, Liu, Y, Hyster, T.K. | | 登録日 | 2023-12-14 | | 公開日 | 2024-03-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Asymmetric Synthesis of alpha-Chloroamides via Photoenzymatic Hydroalkylation of Olefins.
J.Am.Chem.Soc., 146, 2024
|
|
3PMG
 
 | |
6CV2
 
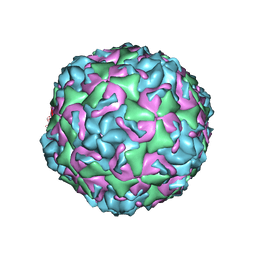 | |
6CVB
 
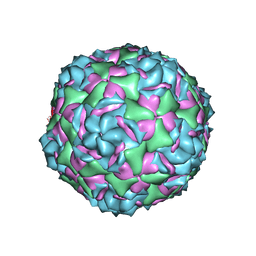 | |
6CV3
 
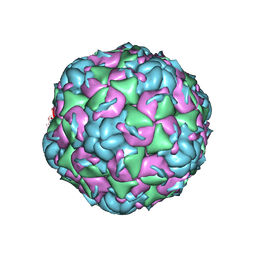 | |
