7Z8E
 
 | | Crystal structure of the substrate-binding protein YejA from S. meliloti in complex with peptide fragment | | 分子名称: | 1,2-ETHANEDIOL, ABC transporter substrate-binding protein, GLY-SER-ASP-VAL-ALA, ... | | 著者 | Morera, S, Vigouroux, V, Travin, D.Y. | | 登録日 | 2022-03-17 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Dual-Uptake Mode of the Antibiotic Phazolicin Prevents Resistance Acquisition by Gram-Negative Bacteria.
Mbio, 14, 2023
|
|
4EUK
 
 | | Crystal structure | | 分子名称: | 1,2-ETHANEDIOL, Histidine kinase 5, Histidine-containing phosphotransfer protein 1, ... | | 著者 | Stehle, T, Bauer, J. | | 登録日 | 2012-04-25 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-Function Analysis of Arabidopsis thaliana Histidine Kinase AHK5 Bound to Its Cognate Phosphotransfer Protein AHP1.
Mol Plant, 6, 2013
|
|
6MME
 
 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | 著者 | Phillips, R.S. | | 登録日 | 2018-09-30 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MQQ
 
 | | Citrobacter freundii F448A mutant tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | 著者 | Phillips, R.S. | | 登録日 | 2018-10-10 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6NV8
 
 | | Perdeuterated tyrosine phenol-lyase from Citrobacter freundii complexed with an aminoacrylate intermediate formed from S-ethyl-L-cysteine and 4-hydroxypyridine | | 分子名称: | 2-AMINO-ACRYLIC ACID, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | 著者 | Phillips, R.S. | | 登録日 | 2019-02-04 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
3DL1
 
 | |
6OYB
 
 | |
6OYE
 
 | |
6OYG
 
 | |
6OY8
 
 | |
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2022-01-03 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7O6N
 
 | |
7OCX
 
 | |
7OCZ
 
 | |
7O6L
 
 | |
7PEB
 
 | |
7PE9
 
 | |
7PEC
 
 | |
7PE8
 
 | |
7PED
 
 | |
7PEA
 
 | |
7PE7
 
 | |
3BDJ
 
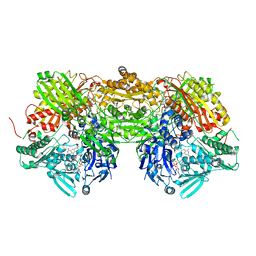 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with a Covalently Bound Oxipurinol Inhibitor | | 分子名称: | CALCIUM ION, CARBONATE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F, Nishino, T. | | 登録日 | 2007-11-14 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanism of inhibition of xanthine oxidoreductase by allopurinol: crystal structure of reduced bovine milk xanthine oxidoreductase bound with oxipurinol.
Nucleosides Nucleotides Nucleic Acids, 27, 2008
|
|
3FL5
 
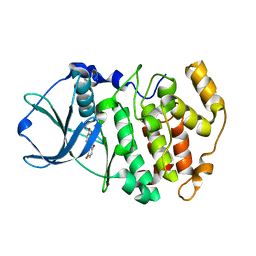 | | Protein kinase CK2 in complex with the inhibitor Quinalizarin | | 分子名称: | 1,2,5,8-tetrahydroxyanthracene-9,10-dione, Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER | | 著者 | Mazzorana, M, Franchin, C, Battistutta, R. | | 登録日 | 2008-12-18 | | 公開日 | 2009-08-18 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Quinalizarin as a potent, selective and cell-permeable inhibitor of protein kinase CK2
Biochem.J., 421, 2009
|
|
7SI9
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-10-12 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
